Water Determines the Structure and Dynamics of Proteins
- PMID: 27186992
- PMCID: PMC7116073
- DOI: 10.1021/acs.chemrev.5b00664
Water Determines the Structure and Dynamics of Proteins
Abstract
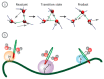
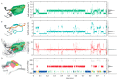
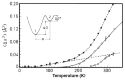
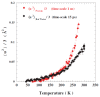
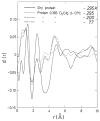
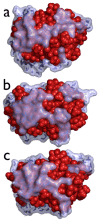
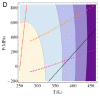
Water is an essential participant in the stability, structure, dynamics, and function of proteins and other biomolecules. Thermodynamically, changes in the aqueous environment affect the stability of biomolecules. Structurally, water participates chemically in the catalytic function of proteins and nucleic acids and physically in the collapse of the protein chain during folding through hydrophobic collapse and mediates binding through the hydrogen bond in complex formation. Water is a partner that slaves the dynamics of proteins, and water interaction with proteins affect their dynamics. Here we provide a review of the experimental and computational advances over the past decade in understanding the role of water in the dynamics, structure, and function of proteins. We focus on the combination of X-ray and neutron crystallography, NMR, terahertz spectroscopy, mass spectroscopy, thermodynamics, and computer simulations to reveal how water assist proteins in their function. The recent advances in computer simulations and the enhanced sensitivity of experimental tools promise major advances in the understanding of protein dynamics, and water surely will be a protagonist.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

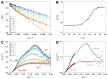
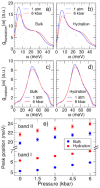
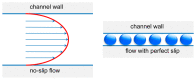
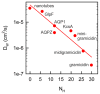
Similar articles
-
Characterization of the denaturation of human alpha-lactalbumin in urea by molecular dynamics simulations.Proteins. 2005 Feb 1;58(2):439-49. doi: 10.1002/prot.20287. Proteins. 2005. PMID: 15558602
-
Protein simulations with an optimized water model: cooperative helix formation and temperature-induced unfolded state collapse.J Phys Chem B. 2010 Nov 25;114(46):14916-23. doi: 10.1021/jp108618d. Epub 2010 Nov 1. J Phys Chem B. 2010. PMID: 21038907
-
Contribution of Water to Pressure and Cold Denaturation of Proteins.Phys Rev Lett. 2015 Sep 4;115(10):108101. doi: 10.1103/PhysRevLett.115.108101. Epub 2015 Sep 1. Phys Rev Lett. 2015. PMID: 26382703
-
Molecular dynamics simulation of proteins under high pressure: Structure, function and thermodynamics.Biochim Biophys Acta Gen Subj. 2020 Feb;1864(2):129395. doi: 10.1016/j.bbagen.2019.07.004. Epub 2019 Jul 11. Biochim Biophys Acta Gen Subj. 2020. PMID: 31302180 Review.
-
Self-similar dynamics of proteins under hydrostatic pressure-Computer simulations and experiments.Biochim Biophys Acta. 2010 Jan;1804(1):56-62. doi: 10.1016/j.bbapap.2009.05.007. Epub 2009 Jun 18. Biochim Biophys Acta. 2010. PMID: 19540369 Review.
Cited by
-
Coupling Monte Carlo, Variational Implicit Solvation, and Binary Level-Set for Simulations of Biomolecular Binding.J Chem Theory Comput. 2021 Apr 13;17(4):2465-2478. doi: 10.1021/acs.jctc.0c01109. Epub 2021 Mar 2. J Chem Theory Comput. 2021. PMID: 33650860 Free PMC article.
-
An integrative structural study of the human full-length RAD52 at 2.2 Å resolution.Commun Biol. 2024 Aug 8;7(1):956. doi: 10.1038/s42003-024-06644-1. Commun Biol. 2024. PMID: 39112549 Free PMC article.
-
Control of Transcription Initiation by Biased Thermal Fluctuations on Repetitive Genomic Sequences.Biomolecules. 2020 Sep 9;10(9):1299. doi: 10.3390/biom10091299. Biomolecules. 2020. PMID: 32916947 Free PMC article.
-
Water-oriented magnetic anisotropy transition.Nat Commun. 2021 May 12;12(1):2738. doi: 10.1038/s41467-021-23057-4. Nat Commun. 2021. PMID: 33980833 Free PMC article.
-
Deciphering Density Fluctuations in the Hydration Water of Brownian Nanoparticles via Upconversion Thermometry.J Phys Chem Lett. 2024 Mar 7;15(9):2606-2615. doi: 10.1021/acs.jpclett.4c00044. Epub 2024 Feb 29. J Phys Chem Lett. 2024. PMID: 38420927 Free PMC article.
References
-
- Kauzmann W. Some Factors in The Interpretation of Protein Denaturation. Adv Protein Chem. 1959;14:1–63. - PubMed
-
- Anfinsen CB. Principles That Govern The Folding of Protein Chains. Science. 1973;181(4096):223–230. - PubMed
-
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular Chaperones in Protein Folding and Proteostasis. Nature. 2011;475(7356):324–332. - PubMed
-
- Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU. Molecular Chaperone Functions in Protein Folding and Proteostasis. Annu Rev Biochem. 2013;82:323–355. - PubMed
-
- Makhatadze GI, Privalov PL. Contribution of Hydration To Protein-Folding Thermodynamics 0.1. The Enthalpy of Hydration. J Mol Biol. 1993;232(2):639–659. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

