Diaphanous formin mDia2 regulates CENP-A levels at centromeres
- PMID: 27185834
- PMCID: PMC4878093
- DOI: 10.1083/jcb.201512034
Diaphanous formin mDia2 regulates CENP-A levels at centromeres
Abstract
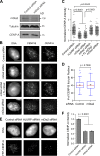
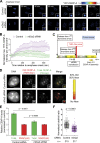
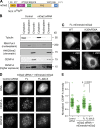
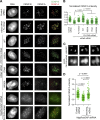
Centromeres of higher eukaryotes are epigenetically defined by centromere protein A (CENP-A), a centromere-specific histone H3 variant. The incorporation of new CENP-A into centromeres to maintain the epigenetic marker after genome replication in S phase occurs in G1 phase; however, how new CENP-A is loaded and stabilized remains poorly understood. Here, we identify the formin mDia2 as essential for stable replenishment of new CENP-A at centromeres. Quantitative imaging, pulse-chase analysis, and high-resolution ratiometric live-cell studies demonstrate that mDia2 and its nuclear localization are required to maintain CENP-A levels at centromeres. Depletion of mDia2 results in a prolonged centromere association of holiday junction recognition protein (HJURP), the chaperone required for CENP-A loading. A constitutively active form of mDia2 rescues the defect in new CENP-A loading caused by depletion of male germ cell Rac GTPase-activating protein (MgcRacGAP), a component of the small GTPase pathway essential for CENP-A maintenance. Thus, the formin mDia2 functions downstream of the MgcRacGAP-dependent pathway in regulating assembly of new CENP-A containing nucleosomes at centromeres.
© 2016 Liu and Mao.
Figures
Similar articles
-
Formin-mediated epigenetic maintenance of centromere identity.Small GTPases. 2017 Oct 2;8(4):245-250. doi: 10.1080/21541248.2016.1215658. Epub 2016 Jul 22. Small GTPases. 2017. PMID: 27449713 Free PMC article. Review.
-
A small GTPase molecular switch regulates epigenetic centromere maintenance by stabilizing newly incorporated CENP-A.Nat Cell Biol. 2010 Dec;12(12):1186-93. doi: 10.1038/ncb2129. Epub 2010 Nov 21. Nat Cell Biol. 2010. PMID: 21102442
-
Nuclear Actin Polymerized by mDia2 Confines Centromere Movement during CENP-A Loading.iScience. 2018 Nov 30;9:314-327. doi: 10.1016/j.isci.2018.10.031. Epub 2018 Nov 2. iScience. 2018. PMID: 30448731 Free PMC article.
-
H3.3 is deposited at centromeres in S phase as a placeholder for newly assembled CENP-A in G₁ phase.Nucleus. 2011 Mar-Apr;2(2):146-57. doi: 10.4161/nucl.2.2.15211. Nucleus. 2011. PMID: 21738837 Free PMC article.
-
Putting CENP-A in its place.Cell Mol Life Sci. 2013 Feb;70(3):387-406. doi: 10.1007/s00018-012-1048-8. Epub 2012 Jun 23. Cell Mol Life Sci. 2013. PMID: 22729156 Free PMC article. Review.
Cited by
-
Role of Anillin in Tumour: From a Prognostic Biomarker to a Novel Target.Cancers (Basel). 2020 Jun 17;12(6):1600. doi: 10.3390/cancers12061600. Cancers (Basel). 2020. PMID: 32560530 Free PMC article. Review.
-
Formin-mediated epigenetic maintenance of centromere identity.Small GTPases. 2017 Oct 2;8(4):245-250. doi: 10.1080/21541248.2016.1215658. Epub 2016 Jul 22. Small GTPases. 2017. PMID: 27449713 Free PMC article. Review.
-
mDia formins form hetero-oligomers and cooperatively maintain murine hematopoiesis.PLoS Genet. 2023 Dec 29;19(12):e1011084. doi: 10.1371/journal.pgen.1011084. eCollection 2023 Dec. PLoS Genet. 2023. PMID: 38157491 Free PMC article.
-
DIAPH1 Promotes Laryngeal Squamous Cell Carcinoma Progression Through Cell Cycle Regulation.Front Oncol. 2021 Sep 22;11:716876. doi: 10.3389/fonc.2021.716876. eCollection 2021. Front Oncol. 2021. PMID: 34631544 Free PMC article.
-
Set Theory, Logic, and Probability: The Integration of Qualitative Reasoning into Teaching Statistics for Quantitative Biology.CBE Life Sci Educ. 2016 winter;15(4):le3. doi: 10.1187/cbe.16-06-0184. CBE Life Sci Educ. 2016. PMID: 27789530 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

