Anatomy of RISC: how do small RNAs and chaperones activate Argonaute proteins?
- PMID: 27184117
- PMCID: PMC5084781
- DOI: 10.1002/wrna.1356
Anatomy of RISC: how do small RNAs and chaperones activate Argonaute proteins?
Abstract
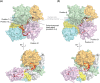
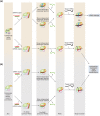
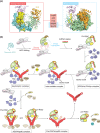
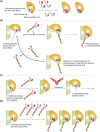
RNA silencing is a eukaryote-specific phenomenon in which microRNAs and small interfering RNAs degrade messenger RNAs containing a complementary sequence. To this end, these small RNAs need to be loaded onto an Argonaute protein (AGO protein) to form the effector complex referred to as RNA-induced silencing complex (RISC). RISC assembly undergoes multiple and sequential steps with the aid of Hsc70/Hsp90 chaperone machinery. The molecular mechanisms for this assembly process remain unclear, despite their significance for the development of gene silencing techniques and RNA interference-based therapeutics. This review dissects the currently available structures of AGO proteins and proposes models and hypotheses for RISC assembly, covering the conformation of unloaded AGO proteins, the chaperone-assisted duplex loading, and the slicer-dependent and slicer-independent duplex separation. The differences in the properties of RISC between prokaryotes and eukaryotes will also be clarified. WIREs RNA 2016, 7:637-660. doi: 10.1002/wrna.1356 For further resources related to this article, please visit the WIREs website.
© 2016 The Authors. WIREs RNA published by Wiley Periodicals, Inc.
Figures



Similar articles
-
Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes.Mol Cell. 2010 Jul 30;39(2):292-9. doi: 10.1016/j.molcel.2010.05.015. Epub 2010 Jun 3. Mol Cell. 2010. PMID: 20605501
-
RISC assembly: Coordination between small RNAs and Argonaute proteins.Biochim Biophys Acta. 2016 Jan;1859(1):71-81. doi: 10.1016/j.bbagrm.2015.08.007. Epub 2015 Aug 22. Biochim Biophys Acta. 2016. PMID: 26303205 Review.
-
The co-chaperones Fkbp4/5 control Argonaute2 expression and facilitate RISC assembly.RNA. 2013 Nov;19(11):1583-93. doi: 10.1261/rna.040790.113. Epub 2013 Sep 18. RNA. 2013. PMID: 24049110 Free PMC article.
-
Reconstitution of RNA Interference Machinery.Methods Mol Biol. 2018;1680:131-143. doi: 10.1007/978-1-4939-7339-2_9. Methods Mol Biol. 2018. PMID: 29030846
-
Life of RISC: Formation, action, and degradation of RNA-induced silencing complex.Mol Cell. 2022 Jan 6;82(1):30-43. doi: 10.1016/j.molcel.2021.11.026. Epub 2021 Dec 22. Mol Cell. 2022. PMID: 34942118 Review.
Cited by
-
RAB18 is a key regulator of GalNAc-conjugated siRNA-induced silencing in Hep3B cells.Mol Ther Nucleic Acids. 2022 Apr 2;28:423-434. doi: 10.1016/j.omtn.2022.04.003. eCollection 2022 Jun 14. Mol Ther Nucleic Acids. 2022. PMID: 35505960 Free PMC article.
-
Exosomes and miRNA-Loaded Biomimetic Nanovehicles, a Focus on Their Potentials Preventing Type-2 Diabetes Linked to Metabolic Syndrome.Front Immunol. 2018 Nov 21;9:2711. doi: 10.3389/fimmu.2018.02711. eCollection 2018. Front Immunol. 2018. PMID: 30519245 Free PMC article. Review.
-
miRNAs in depression vulnerability and resilience: novel targets for preventive strategies.J Neural Transm (Vienna). 2019 Sep;126(9):1241-1258. doi: 10.1007/s00702-019-02048-2. Epub 2019 Jul 26. J Neural Transm (Vienna). 2019. PMID: 31350592 Free PMC article. Review.
-
Regulation of microRNA biogenesis and its crosstalk with other cellular pathways.Nat Rev Mol Cell Biol. 2019 Jan;20(1):5-20. doi: 10.1038/s41580-018-0059-1. Nat Rev Mol Cell Biol. 2019. PMID: 30228348 Review.
-
New opportunities for designing effective small interfering RNAs.Sci Rep. 2019 Nov 6;9(1):16146. doi: 10.1038/s41598-019-52303-5. Sci Rep. 2019. PMID: 31695077 Free PMC article.
References
-
- Meister G. Argonaute proteins: functional insights and emerging roles. Nat Rev Genet 2013, 14:447–459. - PubMed
-
- Kawamata T, Tomari Y. Making RISC. Trends Biochem Sci 2010, 35:368–376. - PubMed
-
- Jinek M, Doudna JA. A three‐dimensional view of the molecular machinery of RNA interference. Nature 2009, 457:405–412. - PubMed
-
- Jonas S, Izaurralde E. Towards a molecular understanding of microRNA‐mediated gene silencing. Nat Rev Genet 2015, 16:421–433. - PubMed
-
- Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet 2011, 12:99–110. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

