Composing a Tumor Specific Bacterial Promoter
- PMID: 27171245
- PMCID: PMC4865170
- DOI: 10.1371/journal.pone.0155338
Composing a Tumor Specific Bacterial Promoter
Abstract
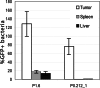
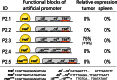
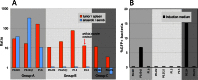
Systemically applied Salmonella enterica spp. have been shown to invade and colonize neoplastic tissues where it retards the growth of many tumors. This offers the possibility to use the bacteria as a vehicle for the tumor specific delivery of therapeutic molecules. Specificity of such delivery is solely depending on promoter sequences that control the production of a target molecule. We have established the functional structure of bacterial promoters that are transcriptionally active exclusively in tumor tissues after systemic application. We observed that the specific transcriptional activation is accomplished by a combination of a weak basal promoter and a strong FNR binding site. This represents a minimal set of control elements required for such activation. In natural promoters, additional DNA remodeling elements are found that alter the level of transcription quantitatively. Inefficiency of the basal promoter ensures the absence of transcription outside tumors. As a proof of concept, we compiled an artificial promoter sequence from individual motifs representing FNR and basal promoter and showed specific activation in a tumor microenvironment. Our results open possibilities for the generation of promoters with an adjusted level of expression of target proteins in particular for applications in bacterial tumor therapy.
Conflict of interest statement
Figures




Similar articles
-
Expression of in vivo-inducible Salmonella enterica promoters during infection of Caenorhabditis elegans.FEMS Microbiol Lett. 2008 Jan;278(2):236-41. doi: 10.1111/j.1574-6968.2007.01001.x. FEMS Microbiol Lett. 2008. PMID: 18096019
-
In vivo gene regulation in Salmonella spp. by a salicylate-dependent control circuit.Nat Methods. 2007 Nov;4(11):937-42. doi: 10.1038/nmeth1107. Epub 2007 Oct 7. Nat Methods. 2007. PMID: 17922017
-
Salmonella promoters preferentially activated inside tumors.Cancer Res. 2008 Jun 15;68(12):4827-32. doi: 10.1158/0008-5472.CAN-08-0552. Cancer Res. 2008. PMID: 18559530
-
The in vivo dissection of direct RFX-target gene promoters in C. elegans reveals a novel cis-regulatory element, the C-box.Dev Biol. 2012 Aug 15;368(2):415-26. doi: 10.1016/j.ydbio.2012.05.033. Epub 2012 Jun 5. Dev Biol. 2012. PMID: 22683808
-
Identification of tumor-specific Salmonella Typhimurium promoters and their regulatory logic.Nucleic Acids Res. 2012 Apr;40(7):2984-94. doi: 10.1093/nar/gkr1041. Epub 2011 Dec 2. Nucleic Acids Res. 2012. PMID: 22140114 Free PMC article.
Cited by
-
Bacterial delivery of the anti-tumor azurin-like protein Laz to glioblastoma cells.AMB Express. 2020 Mar 27;10(1):59. doi: 10.1186/s13568-020-00995-8. AMB Express. 2020. PMID: 32221741 Free PMC article.
-
Engineering the gut microbiota to treat chronic diseases.Appl Microbiol Biotechnol. 2020 Sep;104(18):7657-7671. doi: 10.1007/s00253-020-10771-0. Epub 2020 Jul 21. Appl Microbiol Biotechnol. 2020. PMID: 32696297 Free PMC article. Review.
-
BestCRM: An Exhaustive Search for Optimal Cis-Regulatory Modules in Promoters Accelerated by the Multidimensional Hash Function.Int J Mol Sci. 2024 Feb 5;25(3):1903. doi: 10.3390/ijms25031903. Int J Mol Sci. 2024. PMID: 38339181 Free PMC article.
-
Guidelines on the performance evaluation of motif recognition methods in bioinformatics.Front Genet. 2023 Feb 7;14:1135320. doi: 10.3389/fgene.2023.1135320. eCollection 2023. Front Genet. 2023. PMID: 36824436 Free PMC article. No abstract available.
-
Advances in bacterial cancer therapies using synthetic biology.Curr Opin Syst Biol. 2017 Oct;5:1-8. doi: 10.1016/j.coisb.2017.05.009. Epub 2017 May 23. Curr Opin Syst Biol. 2017. PMID: 29881788 Free PMC article.
References
-
- Thamm DH, Kurzman ID, King I, Li Z, Sznol M, Dubielzig RR, et al. Systemic administration of an attenuated, tumor-targeting Salmonella typhimurium to dogs with spontaneous neoplasia: phase I evaluation. Clin Cancer Res. 2005;11(13):4827–34. Epub 2005/07/08. doi: 11/13/4827 [pii] 10.1158/1078-0432.CCR-04-2510 . - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

