Transposable elements in the mammalian embryo: pioneers surviving through stealth and service
- PMID: 27161170
- PMCID: PMC4862087
- DOI: 10.1186/s13059-016-0965-5
Transposable elements in the mammalian embryo: pioneers surviving through stealth and service
Abstract
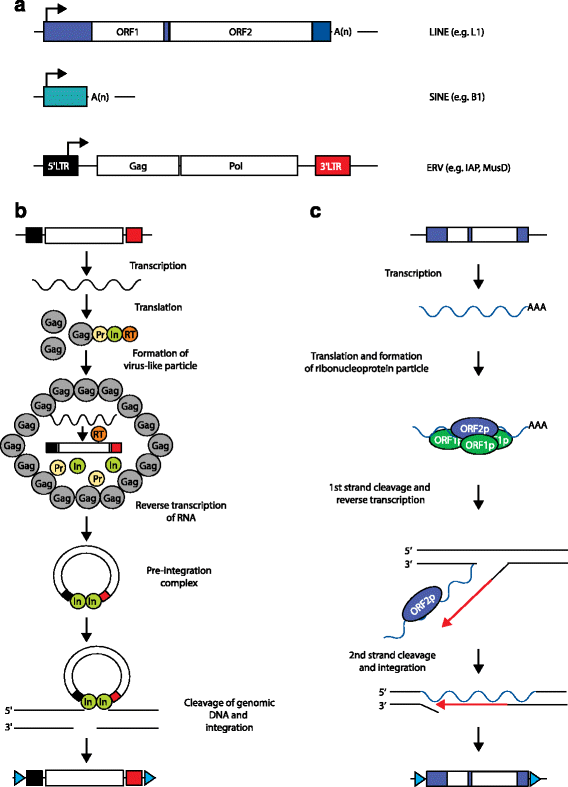
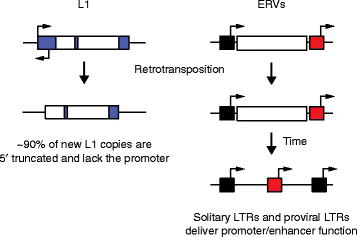
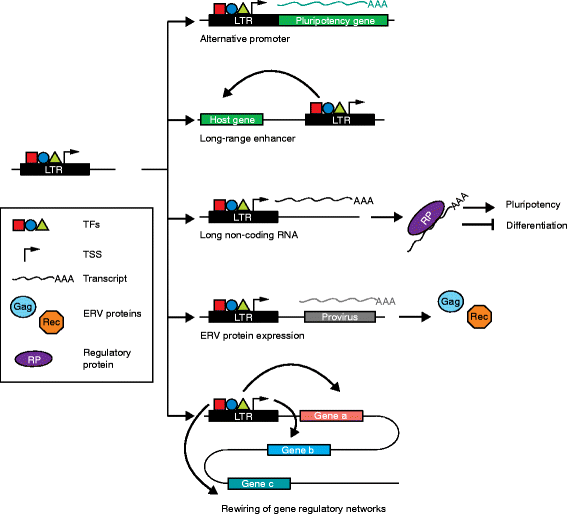
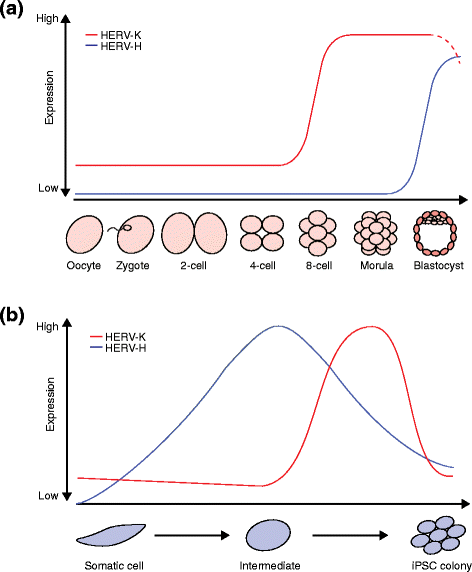
Transposable elements (TEs) are notable drivers of genetic innovation. Over evolutionary time, TE insertions can supply new promoter, enhancer, and insulator elements to protein-coding genes and establish novel, species-specific gene regulatory networks. Conversely, ongoing TE-driven insertional mutagenesis, nonhomologous recombination, and other potentially deleterious processes can cause sporadic disease by disrupting genome integrity or inducing abrupt gene expression changes. Here, we discuss recent evidence suggesting that TEs may contribute regulatory innovation to mammalian embryonic and pluripotent states as a means to ward off complete repression by their host genome.
Figures
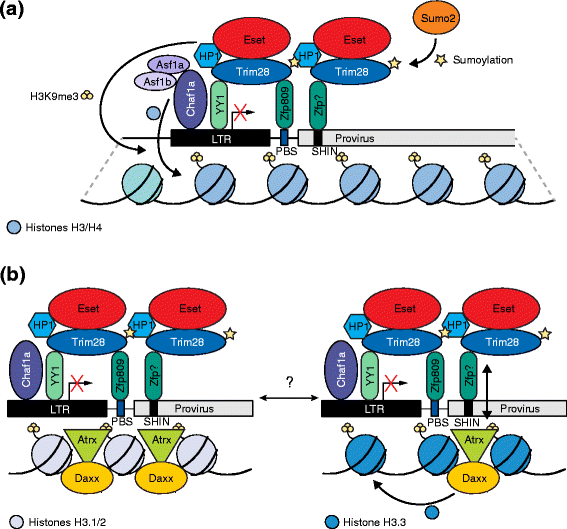
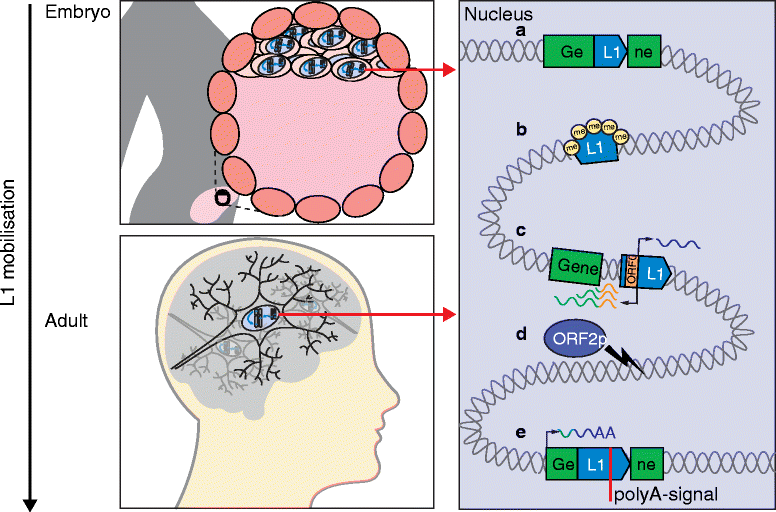
Similar articles
-
The developmental control of transposable elements and the evolution of higher species.Annu Rev Cell Dev Biol. 2015;31:429-51. doi: 10.1146/annurev-cellbio-100814-125514. Epub 2015 Sep 17. Annu Rev Cell Dev Biol. 2015. PMID: 26393776 Review.
-
Functional evaluation of transposable elements as enhancers in mouse embryonic and trophoblast stem cells.Elife. 2019 Apr 23;8:e44344. doi: 10.7554/eLife.44344. Elife. 2019. PMID: 31012843 Free PMC article.
-
Transposable elements as genetic regulatory substrates in early development.Trends Cell Biol. 2013 May;23(5):218-26. doi: 10.1016/j.tcb.2013.01.001. Epub 2013 Feb 12. Trends Cell Biol. 2013. PMID: 23411159 Free PMC article. Review.
-
Host Gene Regulation by Transposable Elements: The New, the Old and the Ugly.Viruses. 2020 Sep 26;12(10):1089. doi: 10.3390/v12101089. Viruses. 2020. PMID: 32993145 Free PMC article. Review.
-
Technology to the rescue: how to uncover the role of transposable elements in preimplantation development.Biochem Soc Trans. 2024 Jun 26;52(3):1349-1362. doi: 10.1042/BST20231262. Biochem Soc Trans. 2024. PMID: 38752836 Free PMC article. Review.
Cited by
-
Trim24 and Trim33 Play a Role in Epigenetic Silencing of Retroviruses in Embryonic Stem Cells.Viruses. 2020 Sep 11;12(9):1015. doi: 10.3390/v12091015. Viruses. 2020. PMID: 32932986 Free PMC article.
-
Transcript assembly improves expression quantification of transposable elements in single-cell RNA-seq data.Genome Res. 2021 Jan;31(1):88-100. doi: 10.1101/gr.265173.120. Epub 2020 Dec 21. Genome Res. 2021. PMID: 33355230 Free PMC article.
-
The coevolution between APOBEC3 and retrotransposons in primates.Mob DNA. 2022 Nov 29;13(1):27. doi: 10.1186/s13100-022-00283-1. Mob DNA. 2022. PMID: 36443831 Free PMC article. Review.
-
CTRL+INSERT: retrotransposons and their contribution to regulation and innovation of the transcriptome.EMBO Rep. 2016 Aug;17(8):1131-44. doi: 10.15252/embr.201642743. Epub 2016 Jul 11. EMBO Rep. 2016. PMID: 27402545 Free PMC article. Review.
-
Reactivation of a somatic errantivirus and germline invasion in Drosophila ovaries.Nat Commun. 2023 Sep 29;14(1):6096. doi: 10.1038/s41467-023-41733-5. Nat Commun. 2023. PMID: 37773253 Free PMC article.
References
-
- Papaioannou VE, Mkandawire J, Biggers JD. Development and phenotypic variability of genetically identical half mouse embryos. Development. 1989;106:817–27. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

