Distinct Gene Regulatory Pathways for Human Innate versus Adaptive Lymphoid Cells
- PMID: 27156452
- PMCID: PMC4874868
- DOI: 10.1016/j.cell.2016.04.014
Distinct Gene Regulatory Pathways for Human Innate versus Adaptive Lymphoid Cells
Abstract
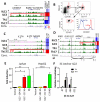
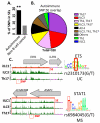
Innate lymphoid cells (ILCs) serve as sentinels in mucosal tissues, sensing release of soluble inflammatory mediators, rapidly communicating danger via cytokine secretion, and functioning as guardians of tissue homeostasis. Although ILCs have been extensively studied in model organisms, little is known about these "first responders" in humans, especially their lineage and functional kinships to cytokine-secreting T helper (Th) cell counterparts. Here, we report gene regulatory circuitries for four human ILC-Th counterparts derived from mucosal environments, revealing that each ILC subset diverges as a distinct lineage from Th and circulating natural killer cells but shares circuitry devoted to functional polarization with their Th counterparts. Super-enhancers demarcate cohorts of cell-identity genes in each lineage, uncovering new modes of regulation for signature cytokines, new molecules that likely impart important functions to ILCs, and potential mechanisms for autoimmune disease SNP associations within ILC-Th subsets.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures




Similar articles
-
Lymphoid tissue inducer-A divergent member of the ILC family.Cytokine Growth Factor Rev. 2018 Aug;42:5-12. doi: 10.1016/j.cytogfr.2018.02.004. Epub 2018 Feb 13. Cytokine Growth Factor Rev. 2018. PMID: 29454785 Free PMC article. Review.
-
Distinct and shared gene expression for human innate versus adaptive helper lymphoid cells.J Leukoc Biol. 2020 Aug;108(2):723-737. doi: 10.1002/JLB.5MA0120-209R. Epub 2020 Feb 4. J Leukoc Biol. 2020. PMID: 32017245 Free PMC article.
-
Multi-Dimensional Gene Regulation in Innate and Adaptive Lymphocytes: A View From Regulomes.Front Immunol. 2021 Mar 25;12:655590. doi: 10.3389/fimmu.2021.655590. eCollection 2021. Front Immunol. 2021. PMID: 33841440 Free PMC article. Review.
-
Transcriptional regulators dictate innate lymphoid cell fates.Protein Cell. 2017 Apr;8(4):242-254. doi: 10.1007/s13238-017-0369-7. Epub 2017 Jan 20. Protein Cell. 2017. PMID: 28108952 Free PMC article. Review.
-
Innate Lymphoid Cells in Mucosal Immunity.Front Immunol. 2019 May 7;10:861. doi: 10.3389/fimmu.2019.00861. eCollection 2019. Front Immunol. 2019. PMID: 31134050 Free PMC article. Review.
Cited by
-
Stage-specific GATA3 induction promotes ILC2 development after lineage commitment.Nat Commun. 2024 Jul 5;15(1):5610. doi: 10.1038/s41467-024-49881-y. Nat Commun. 2024. PMID: 38969652 Free PMC article.
-
Circadian rhythm-dependent and circadian rhythm-independent impacts of the molecular clock on type 3 innate lymphoid cells.Sci Immunol. 2019 Oct 4;4(40):eaay7501. doi: 10.1126/sciimmunol.aay7501. Sci Immunol. 2019. PMID: 31586012 Free PMC article.
-
Innate Lymphoid Cells: Diversity, Plasticity, and Unique Functions in Immunity.Immunity. 2018 Jun 19;48(6):1104-1117. doi: 10.1016/j.immuni.2018.05.013. Immunity. 2018. PMID: 29924976 Free PMC article. Review.
-
Single-cell transcriptomics reveals a senescence-associated IL-6/CCR6 axis driving radiodermatitis.EMBO Mol Med. 2022 Aug 8;14(8):e15653. doi: 10.15252/emmm.202115653. Epub 2022 Jul 4. EMBO Mol Med. 2022. PMID: 35785521 Free PMC article.
-
ILC3s integrate glycolysis and mitochondrial production of reactive oxygen species to fulfill activation demands.J Exp Med. 2019 Oct 7;216(10):2231-2241. doi: 10.1084/jem.20180549. Epub 2019 Jul 11. J Exp Med. 2019. PMID: 31296736 Free PMC article.
References
-
- Ansel KM, Lee DU, Rao A. An epigenetic view of helper T cell differentiation. Nat. Immunol. 2003;4:616–623. - PubMed
-
- Artis D, Spits H. The biology of innate lymphoid cells. Nature. 2015;517:293–301. - PubMed
-
- Bernink JH, Peters CP, Munneke M, te Velde AA, Meijer SL, Weijer K, Hreggvidsdottir HS, Heinsbroek SE, Legrand N, Buskens CJ, et al. Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat. Immunol. 2013;14:221–229. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DE021255/DE/NIDCR NIH HHS/United States
- R21 AI120606/AI/NIAID NIH HHS/United States
- R01 AI118852/AI/NIAID NIH HHS/United States
- R56 AI134035/AI/NIAID NIH HHS/United States
- R37 AI118852/AI/NIAID NIH HHS/United States
- P30 CA091842/CA/NCI NIH HHS/United States
- UL1 TR000448/TR/NCATS NIH HHS/United States
- R21 AI115734/AI/NIAID NIH HHS/United States
- T32 GM007067/GM/NIGMS NIH HHS/United States
- U01 AI095542/AI/NIAID NIH HHS/United States
- R01 CA188286/CA/NCI NIH HHS/United States
- R01 CA176695/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

