Mechanism of Ribonuclease III Catalytic Regulation by Serine Phosphorylation
- PMID: 27150669
- PMCID: PMC4858673
- DOI: 10.1038/srep25448
Mechanism of Ribonuclease III Catalytic Regulation by Serine Phosphorylation
Abstract
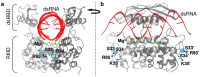
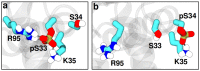
Ribonuclease III (RNase III) is a conserved, gene-regulatory bacterial endonuclease that cleaves double-helical structures in diverse coding and noncoding RNAs. RNase III is subject to multiple levels of control, reflective of its global regulatory functions. Escherichia coli (Ec) RNase III catalytic activity is known to increase during bacteriophage T7 infection, reflecting the expression of the phage-encoded protein kinase, T7PK. However, the mechanism of catalytic enhancement is unknown. This study shows that Ec-RNase III is phosphorylated on serine in vitro by purified T7PK, and identifies the targets as Ser33 and Ser34 in the N-terminal catalytic domain. Kinetic experiments reveal a 5-fold increase in kcat and a 1.4-fold decrease in Km following phosphorylation, providing a 7.4-fold increase in catalytic efficiency. Phosphorylation does not change the rate of substrate cleavage under single-turnover conditions, indicating that phosphorylation enhances product release, which also is the rate-limiting step in the steady-state. Molecular dynamics simulations provide a mechanism for facilitated product release, in which the Ser33 phosphomonoester forms a salt bridge with the Arg95 guanidinium group, thereby weakening RNase III engagement of product. The simulations also show why glutamic acid substitution at either serine does not confer enhancement, thus underscoring the specific requirement for a phosphomonoester.
Figures
Similar articles
-
Mechanism of action of Escherichia coli ribonuclease III. Stringent chemical requirement for the glutamic acid 117 side chain and Mn2+ rescue of the Glu117Asp mutant.Biochemistry. 2001 Apr 24;40(16):5102-10. doi: 10.1021/bi010022d. Biochemistry. 2001. PMID: 11305928
-
Catalytic mechanism of Escherichia coli ribonuclease III: kinetic and inhibitor evidence for the involvement of two magnesium ions in RNA phosphodiester hydrolysis.Nucleic Acids Res. 2005 Feb 7;33(3):807-15. doi: 10.1093/nar/gki197. Print 2005. Nucleic Acids Res. 2005. PMID: 15699182 Free PMC article.
-
Heterodimer-based analysis of subunit and domain contributions to double-stranded RNA processing by Escherichia coli RNase III in vitro.Biochem J. 2008 Feb 15;410(1):39-48. doi: 10.1042/BJ20071047. Biochem J. 2008. PMID: 17953512
-
Regulation of Escherichia coli RNase III activity.J Microbiol. 2015 Aug;53(8):487-94. doi: 10.1007/s12275-015-5323-x. Epub 2015 Jul 31. J Microbiol. 2015. PMID: 26224450 Review.
-
The multifaceted roles of the RNA processing enzyme ribonuclease III.Indian J Biochem Biophys. 1996 Aug;33(4):253-60. Indian J Biochem Biophys. 1996. PMID: 8936814 Review.
Cited by
-
The essential functions of KREPB4 are developmentally distinct and required for endonuclease association with editosomes.RNA. 2017 Nov;23(11):1672-1684. doi: 10.1261/rna.062786.117. Epub 2017 Aug 11. RNA. 2017. PMID: 28802260 Free PMC article.
-
How phosphorylation influences E1 subunit pyruvate dehydrogenase: A computational study.Sci Rep. 2018 Oct 2;8(1):14683. doi: 10.1038/s41598-018-33048-z. Sci Rep. 2018. PMID: 30279533 Free PMC article.
-
Thousands of previously unknown phages discovered in whole-community human gut metagenomes.Microbiome. 2021 Mar 29;9(1):78. doi: 10.1186/s40168-021-01017-w. Microbiome. 2021. PMID: 33781338 Free PMC article.
-
Trans-acting regulators of ribonuclease activity.J Microbiol. 2021 Apr;59(4):341-359. doi: 10.1007/s12275-021-0650-6. Epub 2021 Mar 29. J Microbiol. 2021. PMID: 33779951 Review.
References
-
- Arraiano C. M. et al. The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol. Rev. 34, 883–923 (2010). - PubMed
-
- Evguenieva-Hackenberg E. & Klug G. New aspects of RNA processing in prokaryotes. Curr. Opin. Microbiol. 14, 587–592 (2011). - PubMed
-
- Rochat T., Bouloc P. & Repoila F. Gene expression control by selective RNA processing and stabilization in bacteria. FEMS Microbiol. Lett. 344, 104–103 (2013). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

