The Response of microRNAs to Solar UVR in Skin-Resident Melanocytes Differs between Melanoma Patients and Healthy Persons
- PMID: 27149382
- PMCID: PMC4858311
- DOI: 10.1371/journal.pone.0154915
The Response of microRNAs to Solar UVR in Skin-Resident Melanocytes Differs between Melanoma Patients and Healthy Persons
Abstract
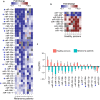
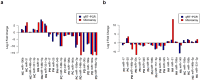
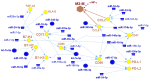
The conversion of melanocytes into cutaneous melanoma is largely dictated by the effects of solar ultraviolet radiation (UVR). Yet to be described, however, is exactly how these cells are affected by intense solar UVR while residing in their natural microenvironment, and whether their response differs in persons with a history of melanoma when compared to that of healthy individuals. By using laser capture microdissection (LCM) to isolate a pure population of melanocytes from a small area of skin that had been intermittingly exposed or un-exposed to physiological doses of solar UVR, we can now report for the first time that the majority of UV-responsive microRNAs (miRNAs) in the melanocytes of a group of women with a history of melanoma are down-regulated when compared to those in the melanocytes of healthy controls. Among the miRNAs that were commonly and significantly down-regulated in each of these women were miR-193b (P<0.003), miR-342-3p (P<0.003), miR186 (P<0.007), miR-130a (P<0.007), and miR-146a (P<0.007). To identify genes potentially released from inhibition by these repressed UV-miRNAs, we analyzed databases (e.g., DIANA-TarBase) containing experimentally validated microRNA-gene interactions. In the end, this enabled us to construct UV-miRNA-gene regulatory networks consisting of individual genes with a probable gain-of-function being intersected not by one, but by several down-regulated UV-miRNAs. Most striking, however, was that these networks typified well-known regulatory modules involved in controlling the epithelial-to-mesenchymal transition and processes associated with the regulation of immune-evasion. We speculate that these pathways become activated by UVR resulting in miRNA down regulation only in melanocytes susceptible to melanoma, and that these changes could be partially responsible for empowering these cells toward tumor progression.
Conflict of interest statement
Figures


Similar articles
-
Premature senescence in human melanocytes after exposure to solar UVR: An exosome and UV-miRNA connection.Pigment Cell Melanoma Res. 2020 Sep;33(5):671-684. doi: 10.1111/pcmr.12888. Epub 2020 Jun 8. Pigment Cell Melanoma Res. 2020. PMID: 32386350
-
UV-Induced Molecular Signaling Differences in Melanoma and Non-melanoma Skin Cancer.Adv Exp Med Biol. 2017;996:27-40. doi: 10.1007/978-3-319-56017-5_3. Adv Exp Med Biol. 2017. PMID: 29124688 Review.
-
Ultraviolet radiation-induced tumor necrosis factor alpha, which is linked to the development of cutaneous SCC, modulates differential epidermal microRNAs expression.Oncotarget. 2016 Apr 5;7(14):17945-56. doi: 10.18632/oncotarget.7595. Oncotarget. 2016. PMID: 26918454 Free PMC article.
-
MicroRNA miR-196a controls melanoma-associated genes by regulating HOX-C8 expression.Int J Cancer. 2011 Sep 1;129(5):1064-74. doi: 10.1002/ijc.25768. Epub 2011 Feb 11. Int J Cancer. 2011. PMID: 21077158
-
Solar ultraviolet-induced DNA damage response: Melanocytes story in transformation to environmental melanomagenesis.Environ Mol Mutagen. 2020 Aug;61(7):736-751. doi: 10.1002/em.22370. Epub 2020 May 8. Environ Mol Mutagen. 2020. PMID: 32281145 Free PMC article. Review.
Cited by
-
Interplay between small and long non-coding RNAs in cutaneous melanoma: a complex jigsaw puzzle with missing pieces.Mol Oncol. 2019 Jan;13(1):74-98. doi: 10.1002/1878-0261.12412. Epub 2018 Dec 20. Mol Oncol. 2019. PMID: 30499222 Free PMC article. Review.
-
Long non-coding RNAs in cutaneous biology and proliferative skin diseases: Advances and perspectives.Cell Prolif. 2020 Jan;53(1):e12698. doi: 10.1111/cpr.12698. Epub 2019 Oct 6. Cell Prolif. 2020. PMID: 31588640 Free PMC article. Review.
-
miRNAs, Melanoma and Microenvironment: An Intricate Network.Int J Mol Sci. 2017 Nov 7;18(11):2354. doi: 10.3390/ijms18112354. Int J Mol Sci. 2017. PMID: 29112174 Free PMC article. Review.
-
Role of non‑coding RNAs in UV‑induced radiation effects (Review).Exp Ther Med. 2024 Apr 23;27(6):262. doi: 10.3892/etm.2024.12550. eCollection 2024 Jun. Exp Ther Med. 2024. PMID: 38756908 Free PMC article. Review.
-
UV-type specific alteration of miRNA expression and its association with tumor progression and metastasis in SCC cell lines.J Cancer Res Clin Oncol. 2020 Dec;146(12):3215-3231. doi: 10.1007/s00432-020-03358-9. Epub 2020 Aug 31. J Cancer Res Clin Oncol. 2020. PMID: 32865618
References
-
- Rastrelli M, Tropea S, Rossi CR, Alaibac M. Melanoma: Epidemiology, Risk Factors, Pathogenesis, Diagnosis and Classification. In vivo. 2014;28(6):1005–11. . - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

