Temporal Dysynchrony in brain connectivity gene expression following hypoxia
- PMID: 27146468
- PMCID: PMC4857255
- DOI: 10.1186/s12864-016-2638-x
Temporal Dysynchrony in brain connectivity gene expression following hypoxia
Abstract
Background: Despite the fundamental biological importance and clinical relevance of characterizing the effects of chronic hypoxia exposure on central nervous system (CNS) development, the changes in gene expression from hypoxia are unknown. It is not known if there are unifying principles, properties, or logic in the response of the developing CNS to hypoxic exposure. Here, we use the small vertebrate zebrafish (Danio rerio) to study the effects of hypoxia on connectivity gene expression across development. We perform transcriptional profiling at high temporal resolution to systematically determine and then experimentally validate the response of CNS connectivity genes to hypoxia exposure.
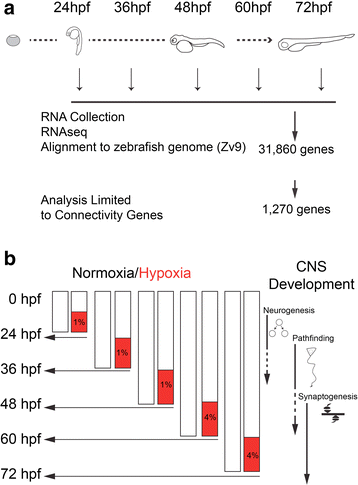
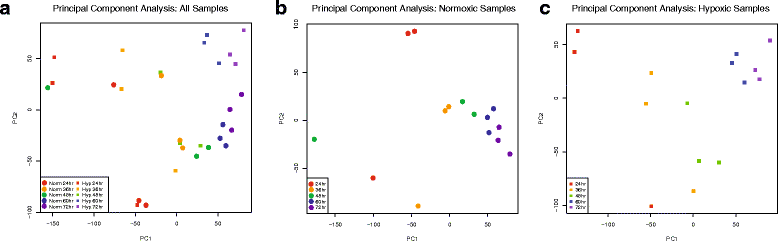
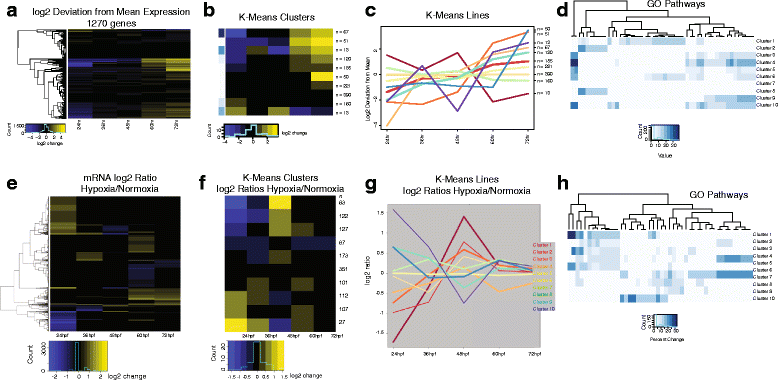
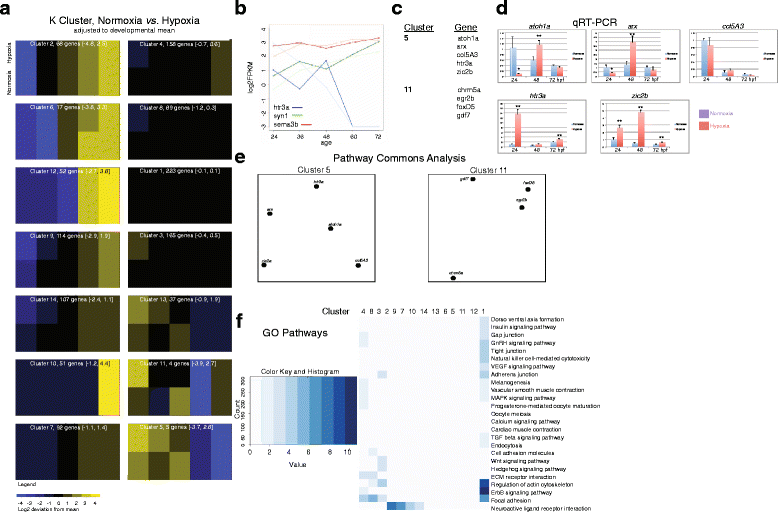
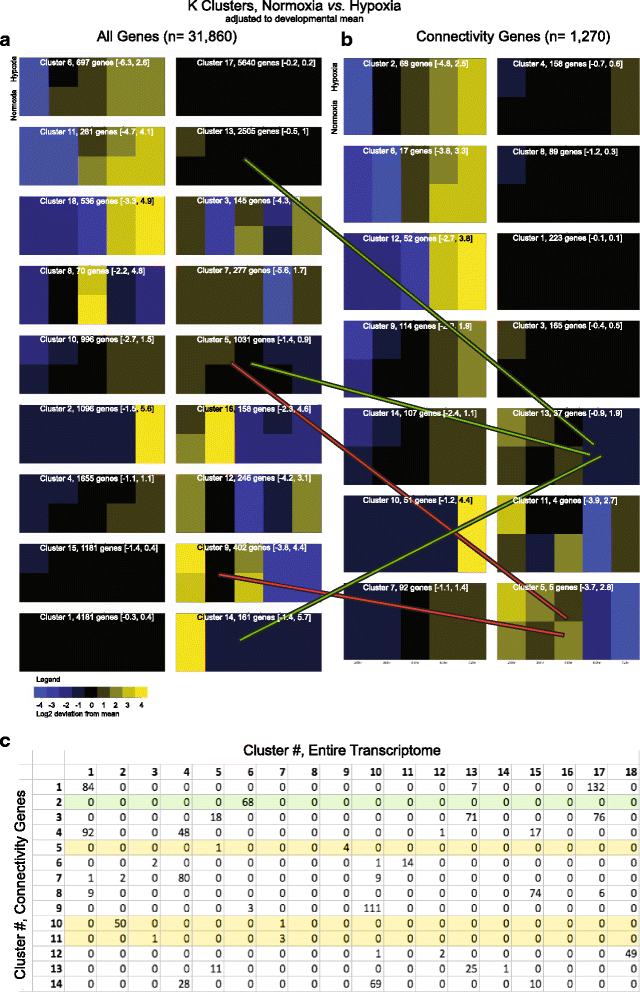
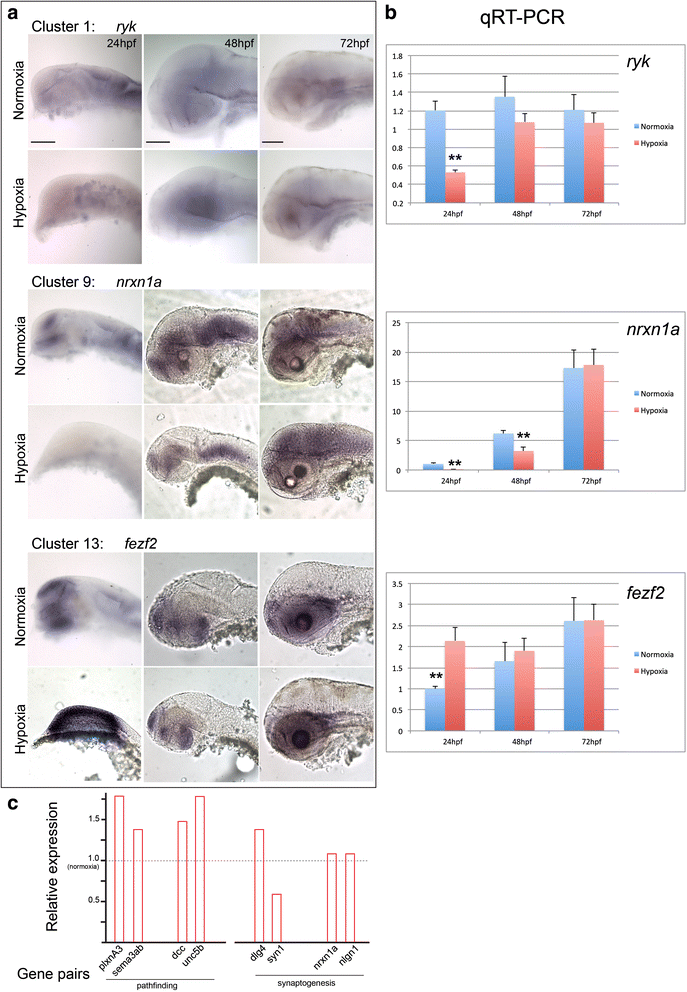
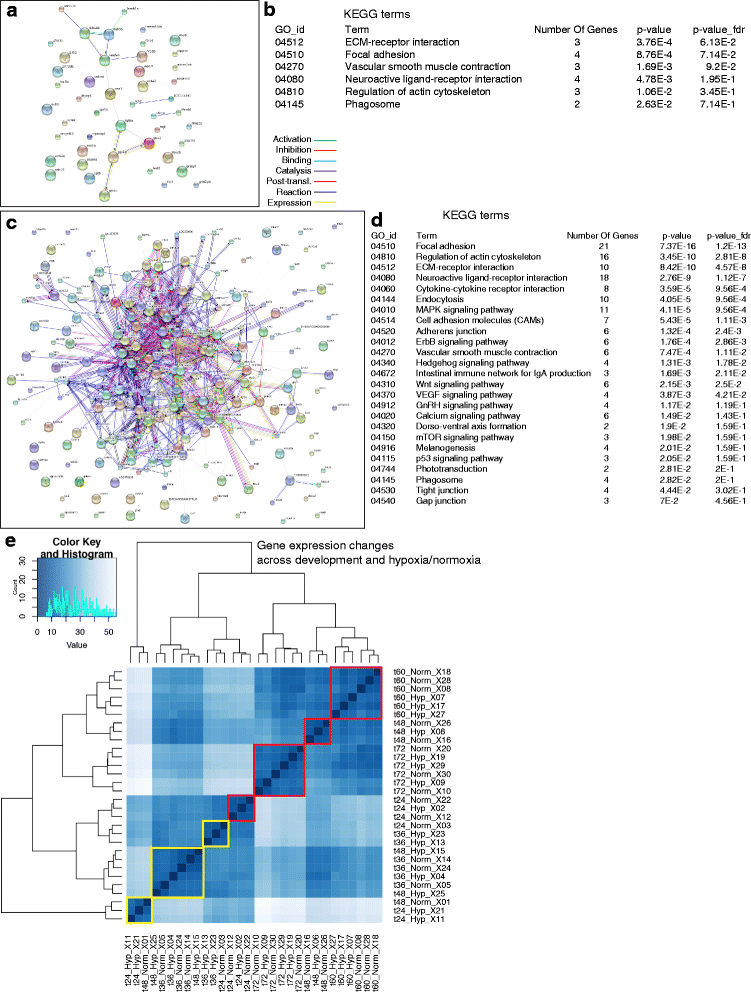
Results: We characterized mRNA changes during development, comparing the effects of chronic hypoxia exposure at different time-points. We focused on changes in expression levels of a subset of 1270 genes selected for their roles in development of CNS connectivity, including axon pathfinding and synapse formation. We found that the majority of CNS connectivity genes were unaffected by hypoxia. However, for a small subset of genes hypoxia significantly affected their gene expression profiles. In particular, hypoxia appeared to affect both the timing and levels of expression, including altering expression of interacting gene pairs in a fashion that would potentially disrupt normal function.
Conclusions: Overall, our study identifies the response of CNS connectivity genes to hypoxia exposure during development. While for most genes hypoxia did not significantly affect expression, for a subset of genes hypoxia changed both levels and timing of expression. Importantly, we identified that some genes with interacting proteins, for example receptor/ligand pairs, had dissimilar responses to hypoxia that would be expected to interfere with their function. The observed dysynchrony of gene expression could impair the development of normal CNS connectivity maps.
Keywords: Axon pathfinding; Connectivity; Hypoxia; Synapse; Zebrafish.
Figures
Similar articles
-
Hypoxia and connectivity in the developing vertebrate nervous system.Dis Model Mech. 2018 Dec 12;11(12):dmm037127. doi: 10.1242/dmm.037127. Dis Model Mech. 2018. PMID: 30541748 Free PMC article. Review.
-
Gene expression profile of zebrafish exposed to hypoxia during development.Physiol Genomics. 2003 Apr 16;13(2):97-106. doi: 10.1152/physiolgenomics.00128.2002. Physiol Genomics. 2003. PMID: 12700360
-
Hypoxia disruption of vertebrate CNS pathfinding through ephrinB2 Is rescued by magnesium.PLoS Genet. 2012;8(4):e1002638. doi: 10.1371/journal.pgen.1002638. Epub 2012 Apr 12. PLoS Genet. 2012. PMID: 22511881 Free PMC article.
-
Cloning of hif-1alpha and hif-2alpha and mRNA expression pattern during development in zebrafish.Gene Expr Patterns. 2007 Jan;7(3):339-45. doi: 10.1016/j.modgep.2006.08.002. Epub 2006 Aug 15. Gene Expr Patterns. 2007. PMID: 16997637
-
Widespread roles of microRNAs during zebrafish development and beyond.Dev Growth Differ. 2012 Jan;54(1):55-65. doi: 10.1111/j.1440-169X.2011.01306.x. Epub 2011 Dec 12. Dev Growth Differ. 2012. PMID: 22150108 Review.
Cited by
-
Dopaminergic Co-Regulation of Locomotor Development and Motor Neuron Synaptogenesis is Uncoupled by Hypoxia in Zebrafish.eNeuro. 2020 Feb 27;7(1):ENEURO.0355-19.2020. doi: 10.1523/ENEURO.0355-19.2020. Print 2020 Jan/Feb. eNeuro. 2020. PMID: 32001551 Free PMC article.
-
Hypoxia and connectivity in the developing vertebrate nervous system.Dis Model Mech. 2018 Dec 12;11(12):dmm037127. doi: 10.1242/dmm.037127. Dis Model Mech. 2018. PMID: 30541748 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

