MiR-211/STAT5A Signaling Modulates Migration of Mesenchymal Stem Cells to Improve its Therapeutic Efficacy
- PMID: 27145179
- PMCID: PMC5096301
- DOI: 10.1002/stem.2391
MiR-211/STAT5A Signaling Modulates Migration of Mesenchymal Stem Cells to Improve its Therapeutic Efficacy
Erratum in
-
Correction to: MiR-211/STAT5A Signaling Modulates Migration of Mesenchymal Stem Cells to Improve its Therapeutic Efficacy.Stem Cells. 2022 Jul 27;40(7):e2. doi: 10.1093/stmcls/sxac028. Stem Cells. 2022. PMID: 35648161 Free PMC article. No abstract available.
Abstract
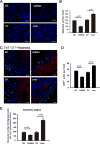
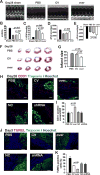
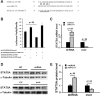
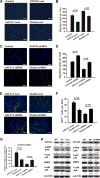
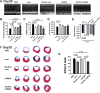
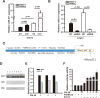
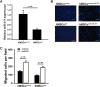
Our previous study showed that the therapeutic effects of mesenchymal stem cells (MSCs) transplantation were improved by enhancing migration. MicroRNA-211 (miR-211) can modulate the migratory properties of some cell types by mechanisms that are not fully understood. This study was designed to investigate a possible role for miR-211 in MSC migration, and whether genetic manipulation of miR-211 in MSCs could be used to enhance its beneficial effects of cell transplantation. Transwell assays confirmed that MSCs migration of was significantly impaired by miR-211 knockdown but enhanced by miR-211 overexpression. MiR-211 overexpressing MSCs also exhibited significantly increased cell engraftment in the peri-infarct areas of female rat hearts 2 days after intravenous transplantation of male MSCs as shown by GFP tracking and SYR gene quantification. This conferred a significant decrease in infarct size and improved cardiac performance. By using a loss or gain of gene function approach, we demonstrated that miR-211 targeted STAT5A to modulate MSCs migration, possibly by interacting with MAPK signaling. Furthermore, the beneficial effects of miR-211 overexpression in MSCs were abolished by simultaneous overexpression of STAT5A whereas the negative effects of miR-211 silencing on MSC migration were rescued by simultaneous downregulation of STAT5A. Finally, using ChIP-PCR and luciferase assays, we provide novel evidence that STAT3 can directly bind to promoter elements that activate miR-211 expression. STAT3/miR-211/STAT5A signaling plays a key role in MSCs migration. Intravenous infusion of genetically modified miR-211 overexpressing MSCs conveys enhanced protection from adverse post-MI remodeling compared with unmodified MSCs. Stem Cells 2016;34:1846-1858.
Keywords: Mesenchymal stem cells; MicroRNA-211; Migration; Myocardial infarction; Retention; STAT5A.
© 2016 The Authors STEM CELLS published by Wiley Periodicals, Inc. on behalf of AlphaMed Press.
Conflict of interest statement
of Potential Conflicts of Interest The authors indicate no potential conflicts of interest.
Figures
Similar articles
-
MicroRNA-133 overexpression promotes the therapeutic efficacy of mesenchymal stem cells on acute myocardial infarction.Stem Cell Res Ther. 2017 Nov 25;8(1):268. doi: 10.1186/s13287-017-0722-z. Stem Cell Res Ther. 2017. PMID: 29178928 Free PMC article.
-
Self-assembling peptide modified with QHREDGS as a novel delivery system for mesenchymal stem cell transplantation after myocardial infarction.FASEB J. 2019 Jul;33(7):8306-8320. doi: 10.1096/fj.201801768RR. Epub 2019 Apr 10. FASEB J. 2019. PMID: 30970221
-
microRNA-206 is involved in survival of hypoxia preconditioned mesenchymal stem cells through targeting Pim-1 kinase.Stem Cell Res Ther. 2016 Apr 22;7(1):61. doi: 10.1186/s13287-016-0318-z. Stem Cell Res Ther. 2016. PMID: 27103465 Free PMC article.
-
MicroRNA-377 regulates mesenchymal stem cell-induced angiogenesis in ischemic hearts by targeting VEGF.PLoS One. 2014 Sep 24;9(9):e104666. doi: 10.1371/journal.pone.0104666. eCollection 2014. PLoS One. 2014. PMID: 25251394 Free PMC article.
-
Follistatin-like 1 protects mesenchymal stem cells from hypoxic damage and enhances their therapeutic efficacy in a mouse myocardial infarction model.Stem Cell Res Ther. 2019 Jan 11;10(1):17. doi: 10.1186/s13287-018-1111-y. Stem Cell Res Ther. 2019. PMID: 30635025 Free PMC article.
Cited by
-
ADSC-derived exosomes attenuate myocardial infarction injury by promoting miR-205-mediated cardiac angiogenesis.Biol Direct. 2023 Feb 27;18(1):6. doi: 10.1186/s13062-023-00361-1. Biol Direct. 2023. PMID: 36849959 Free PMC article.
-
Non-Coding RNAs Steering the Senescence-Related Progress, Properties, and Application of Mesenchymal Stem Cells.Front Cell Dev Biol. 2021 Mar 19;9:650431. doi: 10.3389/fcell.2021.650431. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33816501 Free PMC article. Review.
-
Hepatoma-Derived Growth Factor Secreted from Mesenchymal Stem Cells Reduces Myocardial Ischemia-Reperfusion Injury.Stem Cells Int. 2017;2017:1096980. doi: 10.1155/2017/1096980. Epub 2017 Nov 14. Stem Cells Int. 2017. PMID: 29358952 Free PMC article.
-
Aging of mesenchymal stem cell: machinery, markers, and strategies of fighting.Cell Mol Biol Lett. 2022 Aug 19;27(1):69. doi: 10.1186/s11658-022-00366-0. Cell Mol Biol Lett. 2022. PMID: 35986247 Free PMC article. Review.
-
The Novel miRNA N-72 Regulates EGF-Induced Migration of Human Amnion Mesenchymal Stem Cells by Targeting MMP2.Int J Mol Sci. 2018 May 4;19(5):1363. doi: 10.3390/ijms19051363. Int J Mol Sci. 2018. PMID: 29734654 Free PMC article.
References
-
- Hou D, Youssef EA, Brinton TJ, et al. Radiolabeled cell distribution after intramyocardial, intracoronary, and interstitial retrograde coronary venous delivery: Implications for current clinical trials. Circulation. 2005;112:I150–I156. - PubMed
-
- Bartunek J, Behfar A, Dolatabadi D, et al. Cardiopoietic stem cell therapy in heart failure: The C-CURE (Cardiopoietic stem Cell therapy in heart failURE) multicenter randomized trial with lineage-specified biologics. J Am Coll Cardiol. 2013;61:2329–2338. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

