Identification of genes that are essential to restrict genome duplication to once per cell division
- PMID: 27144335
- PMCID: PMC5085202
- DOI: 10.18632/oncotarget.9008
Identification of genes that are essential to restrict genome duplication to once per cell division
Abstract
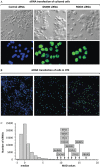
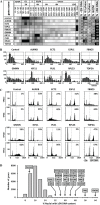
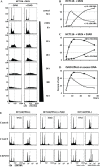
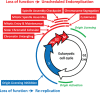
Nuclear genome duplication is normally restricted to once per cell division, but aberrant events that allow excess DNA replication (EDR) promote genomic instability and aneuploidy, both of which are characteristics of cancer development. Here we provide the first comprehensive identification of genes that are essential to restrict genome duplication to once per cell division. An siRNA library of 21,584 human genes was screened for those that prevent EDR in cancer cells with undetectable chromosomal instability. Candidates were validated by testing multiple siRNAs and chemical inhibitors on both TP53+ and TP53- cells to reveal the relevance of this ubiquitous tumor suppressor to preventing EDR, and in the presence of an apoptosis inhibitor to reveal the full extent of EDR. The results revealed 42 genes that prevented either DNA re-replication or unscheduled endoreplication. All of them participate in one or more of eight cell cycle events. Seventeen of them have not been identified previously in this capacity. Remarkably, 14 of the 42 genes have been shown to prevent aneuploidy in mice. Moreover, suppressing a gene that prevents EDR increased the ability of the chemotherapeutic drug Paclitaxel to induce EDR, suggesting new opportunities for synthetic lethalities in the treatment of human cancers.
Keywords: DNA re-replication; aneuploidy; cell cycle regulation; endoreplication; polyploidy.
Conflict of interest statement
The authors have no conflicts of interest.
Figures




Similar articles
-
Genome Duplication at the Beginning of Mammalian Development.Curr Top Dev Biol. 2016;120:55-102. doi: 10.1016/bs.ctdb.2016.04.003. Epub 2016 May 31. Curr Top Dev Biol. 2016. PMID: 27475849 Review.
-
Genome Duplication: The Heartbeat of Developing Organisms.Curr Top Dev Biol. 2016;116:201-29. doi: 10.1016/bs.ctdb.2015.10.002. Epub 2016 Jan 20. Curr Top Dev Biol. 2016. PMID: 26970621 Free PMC article. Review.
-
High-throughput screening for genes that prevent excess DNA replication in human cells and for molecules that inhibit them.Methods. 2012 Jun;57(2):234-48. doi: 10.1016/j.ymeth.2012.03.031. Epub 2012 Apr 5. Methods. 2012. PMID: 22503772 Free PMC article. Review.
-
Guilt by association? p53 and the development of aneuploidy in cancer.Biochem Biophys Res Commun. 2005 Jun 10;331(3):694-700. doi: 10.1016/j.bbrc.2005.03.157. Biochem Biophys Res Commun. 2005. PMID: 15865924 Review.
-
The role of the replication licensing system in cell proliferation and cancer.Prog Cell Cycle Res. 2003;5:287-93. Prog Cell Cycle Res. 2003. PMID: 14593723 Free PMC article. Review.
Cited by
-
Etoposide-induced DNA damage is increased in p53 mutants: identification of ATR and other genes that influence effects of p53 mutations on Top2-induced cytotoxicity.Oncotarget. 2022 Feb 14;13:332-346. doi: 10.18632/oncotarget.28195. eCollection 2022. Oncotarget. 2022. PMID: 35178190 Free PMC article.
-
Non-Canonical Replication Initiation: You're Fired!Genes (Basel). 2017 Jan 27;8(2):54. doi: 10.3390/genes8020054. Genes (Basel). 2017. PMID: 28134821 Free PMC article. Review.
-
Geminin deficiency enhances survival in a murine medulloblastoma model by inducing apoptosis of preneoplastic granule neuron precursors.Genes Cancer. 2017 Sep;8(9-10):725-744. doi: 10.18632/genesandcancer.157. Genes Cancer. 2017. PMID: 29234490 Free PMC article.
-
Adaptive exchange sustains cullin-RING ubiquitin ligase networks and proper licensing of DNA replication.Proc Natl Acad Sci U S A. 2022 Sep 6;119(36):e2205608119. doi: 10.1073/pnas.2205608119. Epub 2022 Aug 29. Proc Natl Acad Sci U S A. 2022. PMID: 36037385 Free PMC article.
-
Validation of Synthetic CRISPR Reagents as a Tool for Arrayed Functional Genomic Screening.PLoS One. 2016 Dec 28;11(12):e0168968. doi: 10.1371/journal.pone.0168968. eCollection 2016. PLoS One. 2016. PMID: 28030641 Free PMC article.
References
-
- Masramon L, Vendrell E, Tarafa G, Capella G, Miro R, Ribas M, Peinado MA. Genetic instability and divergence of clonal populations in colon cancer cells in vitro. J Cell Sci. 2006;119:1477–1482. - PubMed
-
- Geigl JB, Obenauf AC, Schwarzbraun T, Speicher MR. Defining 'chromosomal instability'. Trends in genetics : TIG. 2008;24:64–69. - PubMed
-
- Kuznetsova AY, Seget K, Moeller GK, de Pagter MS, de Roos JA, Durrbaum M, Kuffer C, Muller S, Zaman GJ, Kloosterman WP, Storchova Z. Chromosomal instability, tolerance of mitotic errors and multidrug resistance are promoted by tetraploidization in human cells. Cell Cycle. 2015;14:2810–2820. - PMC - PubMed
-
- Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, Varela I, Phillimore B, Begum S, McDonald NQ, Butler A, Jones D, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. The New England journal of medicine. 2012;366:883–892. - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

