Novel Coronin7 interactions with Cdc42 and N-WASP regulate actin organization and Golgi morphology
- PMID: 27143109
- PMCID: PMC4855144
- DOI: 10.1038/srep25411
Novel Coronin7 interactions with Cdc42 and N-WASP regulate actin organization and Golgi morphology
Abstract
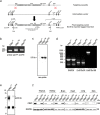
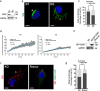
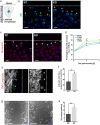
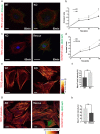
The contribution of the actin cytoskeleton to the unique architecture of the Golgi complex is manifold. An important player in this process is Coronin7 (CRN7), a Golgi-resident protein that stabilizes F-actin assembly at the trans-Golgi network (TGN) thereby facilitating anterograde trafficking. Here, we establish that CRN7-mediated association of F-actin with the Golgi apparatus is distinctly modulated via the small Rho GTPase Cdc42 and N-WASP. We identify N-WASP as a novel interaction partner of CRN7 and demonstrate that CRN7 restricts spurious F-actin reorganizations by repressing N-WASP 'hyperactivity' upon constitutive Cdc42 activation. Loss of CRN7 leads to increased cellular F-actin content and causes a concomitant disruption of the Golgi structure. CRN7 harbours a Cdc42- and Rac-interactive binding (CRIB) motif in its tandem β-propellers and binds selectively to GDP-bound Cdc42N17 mutant. We speculate that CRN7 can act as a cofactor for active Cdc42 generation. Mutation of CRIB motif residues that abrogate Cdc42 binding to CRN7 also fail to rescue the cellular defects in fibroblasts derived from CRN7 KO mice. Cdc42N17 overexpression partially rescued the KO phenotypes whereas N-WASP overexpression failed to do so. We conclude that CRN7 spatiotemporally influences F-actin organization and Golgi integrity in a Cdc42- and N-WASP-dependent manner.
Figures
Similar articles
-
Coronin7 regulates WASP and SCAR through CRIB mediated interaction with Rac proteins.Sci Rep. 2015 Sep 28;5:14437. doi: 10.1038/srep14437. Sci Rep. 2015. PMID: 26411260 Free PMC article.
-
Association of Cdc42/N-WASP/Arp2/3 signaling pathway with Golgi membranes.Traffic. 2004 Nov;5(11):838-46. doi: 10.1111/j.1600-0854.2004.00225.x. Traffic. 2004. PMID: 15479449
-
K33-Linked Polyubiquitination of Coronin 7 by Cul3-KLHL20 Ubiquitin E3 Ligase Regulates Protein Trafficking.Mol Cell. 2014 May 22;54(4):586-600. doi: 10.1016/j.molcel.2014.03.035. Epub 2014 Apr 24. Mol Cell. 2014. PMID: 24768539
-
Cdc42 in actin dynamics: An ordered pathway governed by complex equilibria and directional effector handover.Small GTPases. 2017 Oct 2;8(4):237-244. doi: 10.1080/21541248.2016.1215657. Epub 2016 Aug 11. Small GTPases. 2017. PMID: 27715449 Free PMC article. Review.
-
Regulation of actin dynamics by WASP family proteins.J Biochem. 2003 Sep;134(3):309-13. doi: 10.1093/jb/mvg146. J Biochem. 2003. PMID: 14561714 Review.
Cited by
-
Endogenous and Exogenous Regulatory Signaling in the Secretory Pathway: Role of Golgi Signaling Molecules in Cancer.Front Cell Dev Biol. 2022 Mar 23;10:833663. doi: 10.3389/fcell.2022.833663. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35399533 Free PMC article. Review.
-
FMNL2 and -3 regulate Golgi architecture and anterograde transport downstream of Cdc42.Sci Rep. 2017 Aug 29;7(1):9791. doi: 10.1038/s41598-017-09952-1. Sci Rep. 2017. PMID: 28852060 Free PMC article.
-
Spatiotemporal Control of Intracellular Membrane Trafficking by Rho GTPases.Cells. 2019 Nov 21;8(12):1478. doi: 10.3390/cells8121478. Cells. 2019. PMID: 31766364 Free PMC article. Review.
-
A Proteomic Investigation to Discover Candidate Proteins Involved in Novel Mechanisms of 5-Fluorouracil Resistance in Colorectal Cancer.Cells. 2024 Feb 14;13(4):342. doi: 10.3390/cells13040342. Cells. 2024. PMID: 38391955 Free PMC article.
-
Emerging roles for Rho GTPases operating at the Golgi complex.Small GTPases. 2021 Sep-Nov;12(5-6):311-322. doi: 10.1080/21541248.2020.1812873. Epub 2020 Sep 3. Small GTPases. 2021. PMID: 32881632 Free PMC article. Review.
References
-
- Valderrama F. et al. Actin microfilaments are essential for the cytological positioning and morphology of the Golgi complex. Eur. J. Cell Biol. 76, 9–17 (1998). - PubMed
-
- Etienne-Manneville S. & Hall A. Integrin-mediated activation of Cdc42 controls cell polarity in migrating astrocytes through PKCzeta. Cell 106, 489–98 (2001). - PubMed
-
- Nalbant P., Hodgson L., Kraynov V., Toutchkine A. & Hahn K. M. Activation of endogenous Cdc42 visualized in living cells. Science 305, 1615–1619 (2004). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

