Microsecond Molecular Dynamics Simulations of Influenza Neuraminidase Suggest a Mechanism for the Increased Virulence of Stalk-Deletion Mutants
- PMID: 27141956
- PMCID: PMC5002936
- DOI: 10.1021/acs.jpcb.6b02655
Microsecond Molecular Dynamics Simulations of Influenza Neuraminidase Suggest a Mechanism for the Increased Virulence of Stalk-Deletion Mutants
Abstract
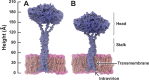
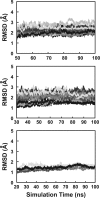
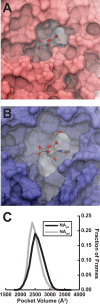
Deletions in the stalk of the influenza neuraminidase (NA) surface protein are associated with increased virulence, but the mechanisms responsible for this enhanced virulence are unclear. Here we use microsecond molecular dynamics simulations to explore the effect of stalk deletion on enzymatic activity, contrasting NA proteins from the A/swine/Shandong/N1/2009 strain both with and without a stalk deletion. By modeling and simulating neuraminidase apo glycoproteins embedded in complex-mixture lipid bilayers, we show that the geometry and dynamics of the neuraminidase enzymatic pocket may differ depending on stalk length, with possible repercussions on the binding of the endogenous sialylated-oligosaccharide receptors. We also use these simulations to predict previously unrecognized druggable "hotspots" on the neuraminidase surface that may prove useful for future efforts aimed at structure-based drug design.
Conflict of interest statement
The authors declare the following competing financial interest(s): R.E.A. is a co-founder of Actavalon, Inc.
Figures


Similar articles
-
An Amino Acid in the Stalk Domain of N1 Neuraminidase Is Critical for Enzymatic Activity.J Virol. 2017 Jan 3;91(2):e00868-16. doi: 10.1128/JVI.00868-16. Print 2017 Jan 15. J Virol. 2017. PMID: 27847354 Free PMC article.
-
[Virological impact of stalk region of neuraminidase in influenza A/Anhui/1/05 (H5N1) and A/Ohio/07/2009 (H1N1) viruses].Bing Du Xue Bao. 2014 May;30(3):238-45. Bing Du Xue Bao. 2014. PMID: 25118377 Chinese.
-
Adaptive mutations of neuraminidase stalk truncation and deglycosylation confer enhanced pathogenicity of influenza A viruses.Sci Rep. 2017 Sep 7;7(1):10928. doi: 10.1038/s41598-017-11348-0. Sci Rep. 2017. PMID: 28883554 Free PMC article.
-
What adaptive changes in hemagglutinin and neuraminidase are necessary for emergence of pandemic influenza virus from its avian precursor?Biochemistry (Mosc). 2015 Jul;80(7):872-80. doi: 10.1134/S000629791507007X. Biochemistry (Mosc). 2015. PMID: 26542001 Review.
-
Efficient incorporation of protein flexibility and dynamics into molecular docking simulations.Biochemistry. 2011 Jul 19;50(28):6157-69. doi: 10.1021/bi2004558. Epub 2011 Jun 22. Biochemistry. 2011. PMID: 21678954 Free PMC article. Review.
Cited by
-
Influenza Virus Neuraminidase Structure and Functions.Front Microbiol. 2019 Jan 29;10:39. doi: 10.3389/fmicb.2019.00039. eCollection 2019. Front Microbiol. 2019. PMID: 30761095 Free PMC article. Review.
-
All-atom virus simulations.Curr Opin Virol. 2018 Aug;31:82-91. doi: 10.1016/j.coviro.2018.08.007. Epub 2018 Sep 1. Curr Opin Virol. 2018. PMID: 30181049 Free PMC article. Review.
-
Building a macro-mixing dual-basin Gō model using the Multistate Bennett Acceptance Ratio.Biophys Physicobiol. 2019 Nov 29;16:310-321. doi: 10.2142/biophysico.16.0_310. eCollection 2019. Biophys Physicobiol. 2019. PMID: 31984186 Free PMC article.
-
Genetic Characterization of Continually Evolving Highly Pathogenic H5N6 Influenza Viruses in China, 2012-2016.Front Microbiol. 2017 Feb 28;8:260. doi: 10.3389/fmicb.2017.00260. eCollection 2017. Front Microbiol. 2017. PMID: 28293218 Free PMC article.
-
Neuraminidase Activity Modulates Cellular Coinfection during Influenza A Virus Multicycle Growth.mBio. 2023 Jun 27;14(3):e0359122. doi: 10.1128/mbio.03591-22. Epub 2023 Apr 20. mBio. 2023. PMID: 37078858 Free PMC article.
References
-
- Cox N. J.; Neumann G.; Donis R. O.; Kawaoka Y.. Orthomyxoviruses: Influenza. In Topley & Wilson’s Microbiology and Microbial Infections; Mahy B. W. J.; Collier L., Eds.; Wiley-Blackwell: London, 2009.
-
- Banks J.; Speidel E. S.; Moore E.; Plowright L.; Piccirillo A.; Capua I.; Cordioli P.; Fioretti A.; Alexander D. J. Changes in the Haemagglutinin and the Neuraminidase Genes Prior to the Emergence of Highly Pathogenic H7n1 Avian Influenza Viruses in Italy. Arch. Virol. 2001, 146, 963–973. 10.1007/s007050170128. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

