Proteomics-driven Antigen Discovery for Development of Vaccines Against Gonorrhea
- PMID: 27141096
- PMCID: PMC4937508
- DOI: 10.1074/mcp.M116.058800
Proteomics-driven Antigen Discovery for Development of Vaccines Against Gonorrhea
Abstract
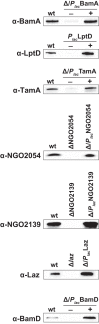
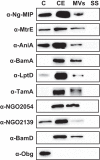
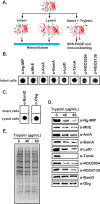
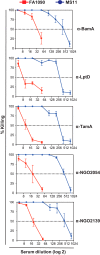
Expanding efforts to develop preventive gonorrhea vaccines is critical because of the dire possibility of untreatable gonococcal infections. Reverse vaccinology, which includes genome and proteome mining, has proven very successful in the discovery of vaccine candidates against many pathogenic bacteria. However, progress with this approach for a gonorrhea vaccine remains in its infancy. Accordingly, we applied a comprehensive proteomic platform-isobaric tagging for absolute quantification coupled with two-dimensional liquid chromatography and mass spectrometry-to identify potential gonococcal vaccine antigens. Our previous analyses focused on cell envelopes and naturally released membrane vesicles derived from four different Neisseria gonorrhoeae strains. Here, we extended these studies to identify cell envelope proteins of N. gonorrhoeae that are ubiquitously expressed and specifically induced by physiologically relevant environmental stimuli: oxygen availability, iron deprivation, and the presence of human serum. Together, these studies enabled the identification of numerous potential gonorrhea vaccine targets. Initial characterization of five novel vaccine candidate antigens that were ubiquitously expressed under these different growth conditions demonstrated that homologs of BamA (NGO1801), LptD (NGO1715), and TamA (NGO1956), and two uncharacterized proteins, NGO2054 and NGO2139, were surface exposed, secreted via naturally released membrane vesicles, and elicited bactericidal antibodies that cross-reacted with a panel of temporally and geographically diverse isolates. In addition, analysis of polymorphisms at the nucleotide and amino acid levels showed that these vaccine candidates are highly conserved among N. gonorrhoeae strains. Finally, depletion of BamA caused a loss of N. gonorrhoeae viability, suggesting it may be an essential target. Together, our data strongly support the use of proteomics-driven discovery of potential vaccine targets as a sound approach for identifying promising gonococcal antigens.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

Similar articles
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Defining the optimum strategy for identifying adults and children with coeliac disease: systematic review and economic modelling.Health Technol Assess. 2022 Oct;26(44):1-310. doi: 10.3310/ZUCE8371. Health Technol Assess. 2022. PMID: 36321689 Free PMC article.
-
Qualitative evidence synthesis informing our understanding of people's perceptions and experiences of targeted digital communication.Cochrane Database Syst Rev. 2019 Oct 23;10(10):ED000141. doi: 10.1002/14651858.ED000141. Cochrane Database Syst Rev. 2019. PMID: 31643081 Free PMC article.
-
Far Posterior Approach for Rib Fracture Fixation: Surgical Technique and Tips.JBJS Essent Surg Tech. 2024 Dec 6;14(4):e23.00094. doi: 10.2106/JBJS.ST.23.00094. eCollection 2024 Oct-Dec. JBJS Essent Surg Tech. 2024. PMID: 39650795 Free PMC article.
-
Conservative, physical and surgical interventions for managing faecal incontinence and constipation in adults with central neurological diseases.Cochrane Database Syst Rev. 2024 Oct 29;10(10):CD002115. doi: 10.1002/14651858.CD002115.pub6. Cochrane Database Syst Rev. 2024. PMID: 39470206
Cited by
-
Sexually Transmitted Neisseria gonorrhoeae Infections-Update on Drug Treatment and Vaccine Development.Medicines (Basel). 2021 Feb 5;8(2):11. doi: 10.3390/medicines8020011. Medicines (Basel). 2021. PMID: 33562607 Free PMC article. Review.
-
Th1-polarized MtrE-based gonococcal vaccines display prophylactic and therapeutic efficacy.Emerg Microbes Infect. 2023 Dec;12(2):2249124. doi: 10.1080/22221751.2023.2249124. Emerg Microbes Infect. 2023. PMID: 37584947 Free PMC article.
-
Dinner date: Neisseria gonorrhoeae central carbon metabolism and pathogenesis.Emerg Top Life Sci. 2024 Feb 22;8(1):15-28. doi: 10.1042/ETLS20220111. Emerg Top Life Sci. 2024. PMID: 37144661 Review.
-
A Borrelia burgdorferi LptD homolog is required for flipping of surface lipoproteins through the spirochetal outer membrane.Mol Microbiol. 2023 Jun;119(6):752-767. doi: 10.1111/mmi.15072. Epub 2023 May 12. Mol Microbiol. 2023. PMID: 37170643 Free PMC article.
-
A Natural Mouse Model for Neisseria Colonization.Infect Immun. 2018 Apr 23;86(5):e00839-17. doi: 10.1128/IAI.00839-17. Print 2018 May. Infect Immun. 2018. PMID: 29440372 Free PMC article.
References
-
- Newman L., Rowley J., Vander Hoorn S., Wijesooriya N. S., Unemo M., Low N., Stevens G., Gottlieb S., Kiarie J., and Temmerman M. (2015) Global estimates of the prevalence and incidence of four curable sexually transmitted infections in 2012 based on systematic review and global reporting. PloS One 10, e0143304. - PMC - PubMed
-
- Lewis D. A. (2010) The Gonococcus fights back: is this time a knock out? Sex. Transm. Infect. 86, 415–421 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

