Dynamic binding of replication protein a is required for DNA repair
- PMID: 27131385
- PMCID: PMC4937323
- DOI: 10.1093/nar/gkw339
Dynamic binding of replication protein a is required for DNA repair
Abstract
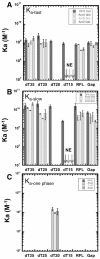
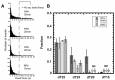
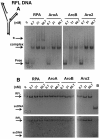
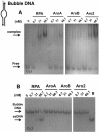
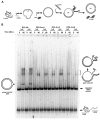
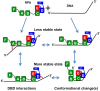
Replication protein A (RPA), the major eukaryotic single-stranded DNA (ssDNA) binding protein, is essential for replication, repair and recombination. High-affinity ssDNA-binding by RPA depends on two DNA binding domains in the large subunit of RPA. Mutation of the evolutionarily conserved aromatic residues in these two domains results in a separation-of-function phenotype: aromatic residue mutants support DNA replication but are defective in DNA repair. We used biochemical and single-molecule analyses, and Brownian Dynamics simulations to determine the molecular basis of this phenotype. Our studies demonstrated that RPA binds to ssDNA in at least two modes characterized by different dissociation kinetics. We also showed that the aromatic residues contribute to the formation of the longer-lived state, are required for stable binding to short ssDNA regions and are needed for RPA melting of partially duplex DNA structures. We conclude that stable binding and/or the melting of secondary DNA structures by RPA is required for DNA repair, including RAD51 mediated DNA strand exchange, but is dispensable for DNA replication. It is likely that the binding modes are in equilibrium and reflect dynamics in the RPA-DNA complex. This suggests that dynamic binding of RPA to DNA is necessary for different cellular functions.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
Concentration-dependent exchange of replication protein A on single-stranded DNA revealed by single-molecule imaging.PLoS One. 2014 Feb 3;9(2):e87922. doi: 10.1371/journal.pone.0087922. eCollection 2014. PLoS One. 2014. PMID: 24498402 Free PMC article.
-
Replication protein A: single-stranded DNA's first responder: dynamic DNA-interactions allow replication protein A to direct single-strand DNA intermediates into different pathways for synthesis or repair.Bioessays. 2014 Dec;36(12):1156-61. doi: 10.1002/bies.201400107. Epub 2014 Aug 29. Bioessays. 2014. PMID: 25171654 Free PMC article. Review.
-
Dynamic regulatory interactions of rad51, rad52, and replication protein-a in recombination intermediates.J Mol Biol. 2009 Jul 3;390(1):45-55. doi: 10.1016/j.jmb.2009.05.009. Epub 2009 May 13. J Mol Biol. 2009. PMID: 19445949
-
Rad52-mediated DNA annealing after Rad51-mediated DNA strand exchange promotes second ssDNA capture.EMBO J. 2006 Nov 29;25(23):5539-48. doi: 10.1038/sj.emboj.7601412. Epub 2006 Nov 9. EMBO J. 2006. PMID: 17093500 Free PMC article.
-
[Replication protein A as a major eukaryotic single-stranded DNA-binding protein and its role in DNA repair].Mol Biol (Mosk). 2016 Sep-Oct;50(5):735-750. doi: 10.7868/S0026898416030083. Mol Biol (Mosk). 2016. PMID: 27830676 Review. Russian.
Cited by
-
Fluorescent human RPA to track assembly dynamics on DNA.Methods. 2024 Mar;223:95-105. doi: 10.1016/j.ymeth.2024.01.019. Epub 2024 Jan 30. Methods. 2024. PMID: 38301751 Free PMC article.
-
RPA and XPA interaction with DNA structures mimicking intermediates of the late stages in nucleotide excision repair.PLoS One. 2018 Jan 10;13(1):e0190782. doi: 10.1371/journal.pone.0190782. eCollection 2018. PLoS One. 2018. PMID: 29320546 Free PMC article.
-
Rad52 Inverse Strand Exchange Drives RNA-Templated DNA Double-Strand Break Repair.Mol Cell. 2017 Jul 6;67(1):19-29.e3. doi: 10.1016/j.molcel.2017.05.019. Epub 2017 Jun 8. Mol Cell. 2017. PMID: 28602639 Free PMC article.
-
The HelQ human DNA repair helicase utilizes a PWI-like domain for DNA loading through interaction with RPA, triggering DNA unwinding by the HelQ helicase core.NAR Cancer. 2021 Jan 12;3(1):zcaa043. doi: 10.1093/narcan/zcaa043. eCollection 2021 Mar. NAR Cancer. 2021. PMID: 34316696 Free PMC article.
-
Dynamic elements of replication protein A at the crossroads of DNA replication, recombination, and repair.Crit Rev Biochem Mol Biol. 2020 Oct;55(5):482-507. doi: 10.1080/10409238.2020.1813070. Epub 2020 Aug 28. Crit Rev Biochem Mol Biol. 2020. PMID: 32856505 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

