Dimerization of SLX4 contributes to functioning of the SLX4-nuclease complex
- PMID: 27131364
- PMCID: PMC4889959
- DOI: 10.1093/nar/gkw354
Dimerization of SLX4 contributes to functioning of the SLX4-nuclease complex
Abstract
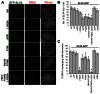
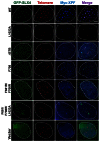
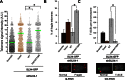
The Fanconi anemia protein SLX4 assembles a genome and telomere maintenance toolkit, consisting of the nucleases SLX1, MUS81 and XPF. Although it is known that SLX4 acts as a scaffold for building this complex, the molecular basis underlying this function of SLX4 remains unclear. Here, we report that functioning of SLX4 is dependent on its dimerization via an oligomerization motif called the BTB domain. We solved the crystal structure of the SLX4BTB dimer, identifying key contacts (F681 and F708) that mediate dimerization. Disruption of BTB dimerization abrogates nuclear foci formation and telomeric localization of not only SLX4 but also of its associated nucleases. Furthermore, dimerization-deficient SLX4 mutants cause defective cellular response to DNA interstrand crosslinking agent and telomere maintenance, underscoring the contribution of BTB domain-mediated dimerization of SLX4 in genome and telomere maintenance.
Published by Oxford University Press on behalf of Nucleic Acids Research 2016. This work is written by (a) US Government employee(s) and is in the public domain in the US.
Figures


Similar articles
-
SLX4 assembles a telomere maintenance toolkit by bridging multiple endonucleases with telomeres.Cell Rep. 2013 Sep 12;4(5):861-9. doi: 10.1016/j.celrep.2013.08.017. Epub 2013 Sep 5. Cell Rep. 2013. PMID: 24012755 Free PMC article.
-
SLX4 contributes to telomere preservation and regulated processing of telomeric joint molecule intermediates.Nucleic Acids Res. 2015 Jul 13;43(12):5912-23. doi: 10.1093/nar/gkv522. Epub 2015 May 18. Nucleic Acids Res. 2015. PMID: 25990736 Free PMC article.
-
Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair.Cell. 2009 Jul 10;138(1):63-77. doi: 10.1016/j.cell.2009.06.030. Cell. 2009. PMID: 19596235 Free PMC article.
-
Structure and mechanism of nucleases regulated by SLX4.Curr Opin Struct Biol. 2016 Feb;36:97-105. doi: 10.1016/j.sbi.2016.01.003. Epub 2016 Jan 29. Curr Opin Struct Biol. 2016. PMID: 26827285 Review.
-
SLX4: multitasking to maintain genome stability.Crit Rev Biochem Mol Biol. 2018 Oct;53(5):475-514. doi: 10.1080/10409238.2018.1488803. Epub 2018 Oct 4. Crit Rev Biochem Mol Biol. 2018. PMID: 30284473 Review.
Cited by
-
Exploring the Structures and Functions of Macromolecular SLX4-Nuclease Complexes in Genome Stability.Front Genet. 2021 Nov 4;12:784167. doi: 10.3389/fgene.2021.784167. eCollection 2021. Front Genet. 2021. PMID: 34804132 Free PMC article. Review.
-
Coordinated roles of SLX4 and MutSβ in DNA repair and the maintenance of genome stability.Crit Rev Biochem Mol Biol. 2021 Apr;56(2):157-177. doi: 10.1080/10409238.2021.1881433. Epub 2021 Feb 17. Crit Rev Biochem Mol Biol. 2021. PMID: 33596761 Free PMC article. Review.
-
In silico analysis of several frequent SLX4 mutations appearing in human cancers.MicroPubl Biol. 2024 May 17;2024:10.17912/micropub.biology.001216. doi: 10.17912/micropub.biology.001216. eCollection 2024. MicroPubl Biol. 2024. PMID: 38828439 Free PMC article.
-
The role of SLX4 and its associated nucleases in DNA interstrand crosslink repair.Nucleic Acids Res. 2019 Mar 18;47(5):2377-2388. doi: 10.1093/nar/gky1276. Nucleic Acids Res. 2019. PMID: 30576517 Free PMC article.
-
Landscape of genetic variants in sporadic meningiomas captured with clinical genomics.Acta Neurochir (Wien). 2022 Sep;164(9):2491-2503. doi: 10.1007/s00701-022-05316-5. Epub 2022 Jul 26. Acta Neurochir (Wien). 2022. PMID: 35881312
References
-
- Stoepker C., Hain K., Schuster B., Hilhorst-Hofstee Y., Rooimans M.A., Steltenpool J., Oostra A.B., Eirich K., Korthof E.T., Nieuwint A.W., et al. SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype. Nat. Genet. 2011;43:138–141. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

