Denoising and covariance estimation of single particle cryo-EM images
- PMID: 27129418
- PMCID: PMC11342156
- DOI: 10.1016/j.jsb.2016.04.013
Denoising and covariance estimation of single particle cryo-EM images
Abstract
The problem of image restoration in cryo-EM entails correcting for the effects of the Contrast Transfer Function (CTF) and noise. Popular methods for image restoration include 'phase flipping', which corrects only for the Fourier phases but not amplitudes, and Wiener filtering, which requires the spectral signal to noise ratio. We propose a new image restoration method which we call 'Covariance Wiener Filtering' (CWF). In CWF, the covariance matrix of the projection images is used within the classical Wiener filtering framework for solving the image restoration deconvolution problem. Our estimation procedure for the covariance matrix is new and successfully corrects for the CTF. We demonstrate the efficacy of CWF by applying it to restore both simulated and experimental cryo-EM images. Results with experimental datasets demonstrate that CWF provides a good way to evaluate the particle images and to see what the dataset contains even without 2D classification and averaging.
Keywords: CTF correction; Steerable PCA; Wiener filtering.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures

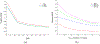
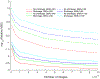






Similar articles
-
Ab-initio contrast estimation and denoising of cryo-EM images.Comput Methods Programs Biomed. 2022 Sep;224:107018. doi: 10.1016/j.cmpb.2022.107018. Epub 2022 Jul 15. Comput Methods Programs Biomed. 2022. PMID: 35901641 Free PMC article.
-
Noise-Transfer2Clean: denoising cryo-EM images based on noise modeling and transfer.Bioinformatics. 2022 Mar 28;38(7):2022-2029. doi: 10.1093/bioinformatics/btac052. Bioinformatics. 2022. PMID: 35134862 Free PMC article.
-
A Fast Image Alignment Approach for 2D Classification of Cryo-EM Images Using Spectral Clustering.Curr Issues Mol Biol. 2021 Oct 18;43(3):1652-1668. doi: 10.3390/cimb43030117. Curr Issues Mol Biol. 2021. PMID: 34698131 Free PMC article.
-
[A review of automatic particle recognition in Cryo-EM images].Sheng Wu Yi Xue Gong Cheng Xue Za Zhi. 2010 Oct;27(5):1178-82. Sheng Wu Yi Xue Gong Cheng Xue Za Zhi. 2010. PMID: 21089695 Review. Chinese.
-
Cryo-electron microscopy for structural biology: current status and future perspectives.Sci China Life Sci. 2015 Aug;58(8):750-6. doi: 10.1007/s11427-015-4851-2. Epub 2015 Apr 19. Sci China Life Sci. 2015. PMID: 25894285 Review.
Cited by
-
Super-resolution multi-reference alignment.Inf inference. 2022 Jun;11(2):533-555. doi: 10.1093/imaiai/iaab003. Epub 2021 Feb 18. Inf inference. 2022. PMID: 35966813 Free PMC article.
-
Self Fourier shell correlation: properties and application to cryo-ET.Commun Biol. 2024 Jan 16;7(1):101. doi: 10.1038/s42003-023-05724-y. Commun Biol. 2024. PMID: 38228756 Free PMC article.
-
Cross-Validation of Data Compatibility Between Small Angle X-ray Scattering and Cryo-Electron Microscopy.J Comput Biol. 2017 Jan;24(1):13-30. doi: 10.1089/cmb.2016.0139. Epub 2016 Oct 6. J Comput Biol. 2017. PMID: 27710115 Free PMC article.
-
Centering Noisy Images with Application to Cryo-EM.SIAM J Imaging Sci. 2021;14(2):689-716. doi: 10.1137/20m1365946. Epub 2021 May 25. SIAM J Imaging Sci. 2021. PMID: 35126803 Free PMC article.
-
Heterogeneous multireference alignment for images with application to 2-D classification in single particle reconstruction.IEEE Trans Image Process. 2019 Oct 10:10.1109/TIP.2019.2945686. doi: 10.1109/TIP.2019.2945686. Online ahead of print. IEEE Trans Image Process. 2019. PMID: 31613760 Free PMC article.
References
-
- Agirrezabala X, Liao HY, Schreiner E, Fu J, Ortiz-Meoz RF, Schulten K, Green R, Frank J, 2012. Structural characterization of mRNA-tRNA translocation intermediates. Proc. Nat. Acad. Sci 109 (16), 6094–6099. 10.1073/pnas.1201288109, arXiv:http://www.pnas.org/content/109/16/6094.full.pdf, URL http://www.pnas.org/content/109/16/6094.abstract. - DOI - PMC - PubMed
-
- Bai XC, McMullan G, Scheres SH, 2015. How cryo-EM is revolutionizing structural biology. Trends Biochem. Sci 40 (1), 49–57. - PubMed
-
- Box GEP, 1976. Science and statistics. J. Am. Stat. Assoc 71 (356), 791–799. 10.1080/01621459.1976.10480949, URL http://www.tandfonline.com/doi/abs/10.1080/01621459.1976.10480949. - DOI - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

