The pluripotency factor Nanog regulates pericentromeric heterochromatin organization in mouse embryonic stem cells
- PMID: 27125671
- PMCID: PMC4863740
- DOI: 10.1101/gad.275685.115
The pluripotency factor Nanog regulates pericentromeric heterochromatin organization in mouse embryonic stem cells
Abstract
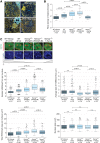
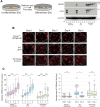
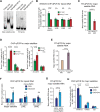
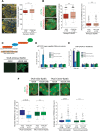
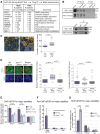
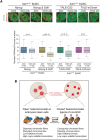
An open and decondensed chromatin organization is a defining property of pluripotency. Several epigenetic regulators have been implicated in maintaining an open chromatin organization, but how these processes are connected to the pluripotency network is unknown. Here, we identified a new role for the transcription factor NANOG as a key regulator connecting the pluripotency network with constitutive heterochromatin organization in mouse embryonic stem cells. Deletion of Nanog leads to chromatin compaction and the remodeling of heterochromatin domains. Forced expression of NANOG in epiblast stem cells is sufficient to decompact chromatin. NANOG associates with satellite repeats within heterochromatin domains, contributing to an architecture characterized by highly dispersed chromatin fibers, low levels of H3K9me3, and high major satellite transcription, and the strong transactivation domain of NANOG is required for this organization. The heterochromatin-associated protein SALL1 is a direct cofactor for NANOG, and loss of Sall1 recapitulates the Nanog-null phenotype, but the loss of Sall1 can be circumvented through direct recruitment of the NANOG transactivation domain to major satellites. These results establish a direct connection between the pluripotency network and chromatin organization and emphasize that maintaining an open heterochromatin architecture is a highly regulated process in embryonic stem cells.
Keywords: embryonic stem cells; heterochromatin; nuclear organization; pluripotency.
© 2016 Novo et al.; Published by Cold Spring Harbor Laboratory Press.
Figures

Similar articles
-
Crosstalk between pluripotency factors and higher-order chromatin organization.Nucleus. 2016 Sep 2;7(5):447-452. doi: 10.1080/19491034.2016.1248013. Nucleus. 2016. PMID: 27759487 Free PMC article. Review.
-
Heterochromatin remodeling in embryonic stem cells proceeds through stochastic de-stabilization of regional steady-states.Biochim Biophys Acta Gene Regul Mech. 2017 Jun;1860(6):661-673. doi: 10.1016/j.bbagrm.2017.01.009. Epub 2017 Jan 20. Biochim Biophys Acta Gene Regul Mech. 2017. PMID: 28115295
-
Pluripotency transcription factors and Tet1/2 maintain Brd4-independent stem cell identity.Nat Cell Biol. 2018 May;20(5):565-574. doi: 10.1038/s41556-018-0086-3. Epub 2018 Apr 16. Nat Cell Biol. 2018. PMID: 29662175 Free PMC article.
-
Chromatin decondensation and nuclear softening accompany Nanog downregulation in embryonic stem cells.Biophys J. 2012 Nov 21;103(10):2060-70. doi: 10.1016/j.bpj.2012.10.015. Epub 2012 Nov 20. Biophys J. 2012. PMID: 23200040 Free PMC article.
-
Phase separation and inheritance of repressive chromatin domains.Curr Opin Genet Dev. 2024 Jun;86:102201. doi: 10.1016/j.gde.2024.102201. Epub 2024 May 2. Curr Opin Genet Dev. 2024. PMID: 38701672 Review.
Cited by
-
Phosphorylation of NANOG by casein kinase I regulates embryonic stem cell self-renewal.FEBS Lett. 2021 Jan;595(1):14-25. doi: 10.1002/1873-3468.13969. Epub 2020 Nov 18. FEBS Lett. 2021. PMID: 33107035 Free PMC article.
-
Derivation of induced pluripotent stem cells from ferret somatic cells.Am J Physiol Lung Cell Mol Physiol. 2020 Apr 1;318(4):L671-L683. doi: 10.1152/ajplung.00456.2019. Epub 2020 Feb 19. Am J Physiol Lung Cell Mol Physiol. 2020. PMID: 32073882 Free PMC article.
-
Tet1 regulates epigenetic remodeling of the pericentromeric heterochromatin and chromocenter organization in DNA hypomethylated cells.PLoS Genet. 2021 Jun 24;17(6):e1009646. doi: 10.1371/journal.pgen.1009646. eCollection 2021 Jun. PLoS Genet. 2021. PMID: 34166371 Free PMC article.
-
A Tale of Two States: Pluripotency Regulation of Telomeres.Front Cell Dev Biol. 2021 Jul 8;9:703466. doi: 10.3389/fcell.2021.703466. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34307383 Free PMC article. Review.
-
Complementary Activity of ETV5, RBPJ, and TCF3 Drives Formative Transition from Naive Pluripotency.Cell Stem Cell. 2019 May 2;24(5):785-801.e7. doi: 10.1016/j.stem.2019.03.017. Epub 2019 Apr 25. Cell Stem Cell. 2019. PMID: 31031137 Free PMC article.
References
-
- Bickmore WA, van Steensel B. 2013. Genome architecture: domain organization of interphase chromosomes. Cell 152: 1270–1284. - PubMed
-
- Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, et al. 2007. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448: 191–195. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
