Revelation of Influencing Factors in Overall Codon Usage Bias of Equine Influenza Viruses
- PMID: 27119730
- PMCID: PMC4847779
- DOI: 10.1371/journal.pone.0154376
Revelation of Influencing Factors in Overall Codon Usage Bias of Equine Influenza Viruses
Abstract
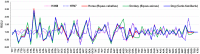
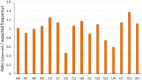
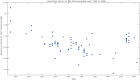
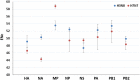
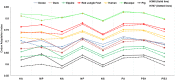
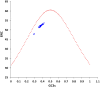
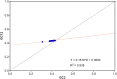
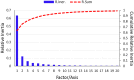
Equine influenza viruses (EIVs) of H3N8 subtype are culprits of severe acute respiratory infections in horses, and are still responsible for significant outbreaks worldwide. Adaptability of influenza viruses to a particular host is significantly influenced by their codon usage preference, due to an absolute dependence on the host cellular machinery for their replication. In the present study, we analyzed genome-wide codon usage patterns in 92 EIV strains, including both H3N8 and H7N7 subtypes by computing several codon usage indices and applying multivariate statistical methods. Relative synonymous codon usage (RSCU) analysis disclosed bias of preferred synonymous codons towards A/U-ended codons. The overall codon usage bias in EIVs was slightly lower, and mainly affected by the nucleotide compositional constraints as inferred from the RSCU and effective number of codon (ENc) analysis. Our data suggested that codon usage pattern in EIVs is governed by the interplay of mutation pressure, natural selection from its hosts and undefined factors. The H7N7 subtype was found less fit to its host (horse) in comparison to H3N8, by possessing higher codon bias, lower mutation pressure and much less adaptation to tRNA pool of equine cells. To the best of our knowledge, this is the first report describing the codon usage analysis of the complete genomes of EIVs. The outcome of our study is likely to enhance our understanding of factors involved in viral adaptation, evolution, and fitness towards their hosts.
Conflict of interest statement
Figures
Similar articles
-
Genetic analysis of the PB1-F2 gene of equine influenza virus.Virus Genes. 2013 Oct;47(2):250-8. doi: 10.1007/s11262-013-0935-x. Epub 2013 Jun 19. Virus Genes. 2013. PMID: 23780220
-
Genetic and codon usage bias analyses of polymerase genes of equine influenza virus and its relation to evolution.BMC Genomics. 2017 Aug 23;18(1):652. doi: 10.1186/s12864-017-4063-1. BMC Genomics. 2017. PMID: 28830350 Free PMC article.
-
Equine Influenza Virus in Asia: Phylogeographic Pattern and Molecular Features Reveal Circulation of an Autochthonous Lineage.J Virol. 2019 Jun 14;93(13):e00116-19. doi: 10.1128/JVI.00116-19. Print 2019 Jul 1. J Virol. 2019. PMID: 31019053 Free PMC article.
-
Equine Influenza Virus and Vaccines.Viruses. 2021 Aug 20;13(8):1657. doi: 10.3390/v13081657. Viruses. 2021. PMID: 34452521 Free PMC article. Review.
-
A Brief Introduction to Equine Influenza and Equine Influenza Viruses.Methods Mol Biol. 2020;2123:355-360. doi: 10.1007/978-1-0716-0346-8_26. Methods Mol Biol. 2020. PMID: 32170701 Review.
Cited by
-
Constraints of Viral RNA Synthesis on Codon Usage of Negative-Strand RNA Virus.J Virol. 2019 Feb 19;93(5):e01775-18. doi: 10.1128/JVI.01775-18. Print 2019 Mar 1. J Virol. 2019. PMID: 30541832 Free PMC article.
-
Comprehensive codon usage analysis of porcine deltacoronavirus.Mol Phylogenet Evol. 2019 Dec;141:106618. doi: 10.1016/j.ympev.2019.106618. Epub 2019 Sep 16. Mol Phylogenet Evol. 2019. PMID: 31536759 Free PMC article.
-
Novel Approaches for The Development of Live Attenuated Influenza Vaccines.Viruses. 2019 Feb 22;11(2):190. doi: 10.3390/v11020190. Viruses. 2019. PMID: 30813325 Free PMC article. Review.
-
Analysis of Nipah Virus Codon Usage and Adaptation to Hosts.Front Microbiol. 2019 May 8;10:886. doi: 10.3389/fmicb.2019.00886. eCollection 2019. Front Microbiol. 2019. PMID: 31156564 Free PMC article.
-
Genomic analysis of codon usage shows influence of mutation pressure, natural selection, and host features on Senecavirus A evolution.Microb Pathog. 2017 Nov;112:313-319. doi: 10.1016/j.micpath.2017.09.040. Epub 2017 Sep 22. Microb Pathog. 2017. PMID: 28943149 Free PMC article.
References
-
- Chen W, Calvo PA, Malide D, Gibbs J, Schubert U, Bacik I, et al. A novel influenza A virus mitochondrial protein that induces cell death. Nat Med. 2001; 7: 1306–1312. - PubMed
-
- Webster RG. Are equine 1 influenza viruses still present in horses? Equine Vet J. 1993; 25: 537–538. - PubMed
-
- Webster RG, Laver WG, Air GM, Schild GC. Molecular mechanisms of variation in influenza viruses. Nature. 1982; 296(5853): 115–121. - PubMed
-
- Hillerman MR. Realities and enigmas of human viral influenza: pathogenesis, epidemiology and control. Vaccine. 2002; 20: 3068–3087. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

