Genome-Wide Chromatin Immunoprecipitation Sequencing Analysis Shows that WhiB Is a Transcription Factor That Cocontrols Its Regulon with WhiA To Initiate Developmental Cell Division in Streptomyces
- PMID: 27094333
- PMCID: PMC4850268
- DOI: 10.1128/mBio.00523-16
Genome-Wide Chromatin Immunoprecipitation Sequencing Analysis Shows that WhiB Is a Transcription Factor That Cocontrols Its Regulon with WhiA To Initiate Developmental Cell Division in Streptomyces
Abstract
WhiB is the founding member of a family of proteins (the WhiB-like [Wbl] family) that carry a [4Fe-4S] iron-sulfur cluster and play key roles in diverse aspects of the biology of actinomycetes, including pathogenesis, antibiotic resistance, and the control of development. In Streptomyces, WhiB is essential for the process of developmentally controlled cell division that leads to sporulation. The biochemical function of Wbl proteins has been controversial; here, we set out to determine unambiguously if WhiB functions as a transcription factor using chromatin immunoprecipitation sequencing (ChIP-seq) in Streptomyces venezuelae. In the first demonstration of in vivo genome-wide Wbl binding, we showed that WhiB regulates the expression of key genes required for sporulation by binding upstream of ~240 transcription units. Strikingly, the WhiB regulon is identical to the previously characterized WhiA regulon, providing an explanation for the identical phenotypes of whiA and whiB mutants. Using ChIP-seq, we demonstrated that in vivo DNA binding by WhiA depends on WhiB and vice versa, showing that WhiA and WhiB function cooperatively to control expression of a common set of WhiAB target genes. Finally, we show that mutation of the cysteine residues that coordinate the [4Fe-4S] cluster in WhiB prevents DNA binding by both WhiB and WhiA in vivo.
Importance: Despite the central importance of WhiB-like (Wbl) proteins in actinomycete biology, a conclusive demonstration of their biochemical function has been elusive, and they have been difficult to study, particularly in vitro, largely because they carry an oxygen-sensitive [4Fe-4S] cluster. Here we used genome-wide ChIP-seq to investigate the function of Streptomyces WhiB, the founding member of the Wbl family. The advantage of this approach is that the oxygen sensitivity of the [4Fe-4S] cluster becomes irrelevant once the protein has been cross-linked to DNA in vivo. Our data provide the most compelling in vivo evidence to date that WhiB, and, by extension, probably all Wbl proteins, function as transcription factors. Further, we show that WhiB does not act independently but rather coregulates its regulon of sporulation genes with a partner transcription factor, WhiA.
Copyright © 2016 Bush et al.
Figures

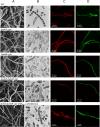
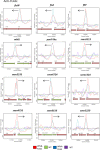
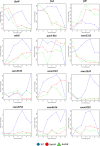
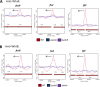

Similar articles
-
The actinobacterial WhiB-like (Wbl) family of transcription factors.Mol Microbiol. 2018 Dec;110(5):663-676. doi: 10.1111/mmi.14117. Epub 2018 Oct 25. Mol Microbiol. 2018. PMID: 30179278 Free PMC article. Review.
-
Genes required for aerial growth, cell division, and chromosome segregation are targets of WhiA before sporulation in Streptomyces venezuelae.mBio. 2013 Sep 24;4(5):e00684-13. doi: 10.1128/mBio.00684-13. mBio. 2013. PMID: 24065632 Free PMC article.
-
The Streptomyces O-B one connection: a force within layered repression of a key developmental decision.Mol Microbiol. 2017 Jun;104(5):695-699. doi: 10.1111/mmi.13688. Epub 2017 Apr 27. Mol Microbiol. 2017. PMID: 28387974
-
Structural basis of dual activation of cell division by the actinobacterial transcription factors WhiA and WhiB.Proc Natl Acad Sci U S A. 2023 Mar 14;120(11):e2220785120. doi: 10.1073/pnas.2220785120. Epub 2023 Mar 8. Proc Natl Acad Sci U S A. 2023. PMID: 36888660 Free PMC article.
-
WhiB-like proteins: Diversity of structure, function and mechanism.Biochim Biophys Acta Mol Cell Res. 2024 Oct;1871(7):119787. doi: 10.1016/j.bbamcr.2024.119787. Epub 2024 Jun 13. Biochim Biophys Acta Mol Cell Res. 2024. PMID: 38879133 Review.
Cited by
-
The actinobacterial WhiB-like (Wbl) family of transcription factors.Mol Microbiol. 2018 Dec;110(5):663-676. doi: 10.1111/mmi.14117. Epub 2018 Oct 25. Mol Microbiol. 2018. PMID: 30179278 Free PMC article. Review.
-
System-level understanding of gene expression and regulation for engineering secondary metabolite production in Streptomyces.J Ind Microbiol Biotechnol. 2020 Oct;47(9-10):739-752. doi: 10.1007/s10295-020-02298-0. Epub 2020 Aug 10. J Ind Microbiol Biotechnol. 2020. PMID: 32778981 Review.
-
Characterization of the Widely Distributed Novel ECF42 Group of Extracytoplasmic Function σ Factors in Streptomyces venezuelae.J Bacteriol. 2018 Oct 10;200(21):e00437-18. doi: 10.1128/JB.00437-18. Print 2018 Nov 1. J Bacteriol. 2018. PMID: 30126941 Free PMC article.
-
Comparative Genomic Insights into Secondary Metabolism Biosynthetic Gene Cluster Distributions of Marine Streptomyces.Mar Drugs. 2019 Aug 26;17(9):498. doi: 10.3390/md17090498. Mar Drugs. 2019. PMID: 31454987 Free PMC article.
-
Multi-layered inhibition of Streptomyces development: BldO is a dedicated repressor of whiB.Mol Microbiol. 2017 Jun;104(5):700-711. doi: 10.1111/mmi.13663. Epub 2017 Mar 27. Mol Microbiol. 2017. PMID: 28271577 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- BBS/E/J/00000015/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/I00873X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/J004561/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/H006125/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

