Metataxonomic and Metagenomic Approaches vs. Culture-Based Techniques for Clinical Pathology
- PMID: 27092134
- PMCID: PMC4823605
- DOI: 10.3389/fmicb.2016.00484
Metataxonomic and Metagenomic Approaches vs. Culture-Based Techniques for Clinical Pathology
Abstract
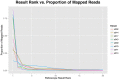
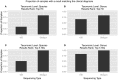
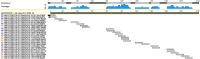
Diagnoses that are both timely and accurate are critically important for patients with life-threatening or drug resistant infections. Technological improvements in High-Throughput Sequencing (HTS) have led to its use in pathogen detection and its application in clinical diagnoses of infectious diseases. The present study compares two HTS methods, 16S rRNA marker gene sequencing (metataxonomics) and whole metagenomic shotgun sequencing (metagenomics), in their respective abilities to match the same diagnosis as traditional culture methods (culture inference) for patients with ventilator associated pneumonia (VAP). The metagenomic analysis was able to produce the same diagnosis as culture methods at the species-level for five of the six samples, while the metataxonomic analysis was only able to produce results with the same species-level identification as culture for two of the six samples. These results indicate that metagenomic analyses have the accuracy needed for a clinical diagnostic tool, but full integration in diagnostic protocols is contingent on technological improvements to decrease turnaround time and lower costs.
Keywords: drug resistance; high throughput sequencing; metagenomics; metataxonomics; microbiome; pathogen detection.
Figures
Similar articles
-
Comparative Analysis of Metagenomics and Metataxonomics for the Characterization of Vermicompost Microbiomes.Front Microbiol. 2022 May 10;13:854423. doi: 10.3389/fmicb.2022.854423. eCollection 2022. Front Microbiol. 2022. PMID: 35620097 Free PMC article.
-
16S rRNA gene sequencing is a non-culture method of defining the specific bacterial etiology of ventilator-associated pneumonia.Int J Clin Exp Med. 2015 Oct 15;8(10):18560-70. eCollection 2015. Int J Clin Exp Med. 2015. PMID: 26770469 Free PMC article.
-
Validation and Retrospective Clinical Evaluation of a Quantitative 16S rRNA Gene Metagenomic Sequencing Assay for Bacterial Pathogen Detection in Body Fluids.J Mol Diagn. 2019 Sep;21(5):913-923. doi: 10.1016/j.jmoldx.2019.05.002. Epub 2019 Jun 21. J Mol Diagn. 2019. PMID: 31229651
-
Metagenomics in pediatrics: using a shotgun approach to diagnose infections.Curr Opin Pediatr. 2018 Feb;30(1):125-130. doi: 10.1097/MOP.0000000000000577. Curr Opin Pediatr. 2018. PMID: 29189352 Review.
-
Clinical Metagenomic Next-Generation Sequencing for Pathogen Detection.Annu Rev Pathol. 2019 Jan 24;14:319-338. doi: 10.1146/annurev-pathmechdis-012418-012751. Epub 2018 Oct 24. Annu Rev Pathol. 2019. PMID: 30355154 Free PMC article. Review.
Cited by
-
Fecal Transplant in Children With Clostridioides difficile Gives Sustained Reduction in Antimicrobial Resistance and Potential Pathogen Burden.Open Forum Infect Dis. 2019 Aug 26;6(10):ofz379. doi: 10.1093/ofid/ofz379. eCollection 2019 Oct. Open Forum Infect Dis. 2019. PMID: 31660343 Free PMC article.
-
The central and biodynamic role of gut microbiota in critically ill patients.Crit Care. 2022 Aug 18;26(1):250. doi: 10.1186/s13054-022-04127-5. Crit Care. 2022. PMID: 35982499 Free PMC article. Review.
-
Metabarcoding of Parasitic Wasp, Dolichogenidea metesae (Nixon) (Hymenoptera: Braconidae) That Parasitizing Bagworm, Metisa plana Walker (Lepidoptera: Psychidae).Trop Life Sci Res. 2022 Mar;33(1):23-42. doi: 10.21315/tlsr2022.33.1.2. Epub 2022 Mar 31. Trop Life Sci Res. 2022. PMID: 35651643 Free PMC article.
-
Gut microbiota from persons with attention-deficit/hyperactivity disorder affects the brain in mice.Microbiome. 2020 Apr 1;8(1):44. doi: 10.1186/s40168-020-00816-x. Microbiome. 2020. PMID: 32238191 Free PMC article.
-
Detection of Pulmonary Infectious Pathogens From Lung Biopsy Tissues by Metagenomic Next-Generation Sequencing.Front Cell Infect Microbiol. 2018 Jun 25;8:205. doi: 10.3389/fcimb.2018.00205. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 29988504 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

