Cellular uptake of exogenous calcineurin B is dependent on TLR4/MD2/CD14 complexes, and CnB is an endogenous ligand of TLR4
- PMID: 27090571
- PMCID: PMC4835703
- DOI: 10.1038/srep24346
Cellular uptake of exogenous calcineurin B is dependent on TLR4/MD2/CD14 complexes, and CnB is an endogenous ligand of TLR4
Abstract
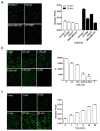
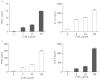
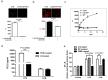
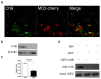
Our previous research showed that recombinant calcineurin B (rhCnB) stimulates cytokine secretion by immune cells, probably through TLR4. Exogenous CnB can be incorporated into many different tumour cells in vitro, but the mode of uptake and receptors required remain unknown. Here, we report that exogenous CnB is taken up by cells in a time- and concentration-dependent manner via clathrin-dependent receptor-mediated internalization. Our findings further confirm that uptake is mediated by the TLR4/MD2 complex together with the co-receptor CD14. The MST results revealed a high affinity between CnB and the TLR4 receptor complex. No binding was detected between CnB and LPS. CnB inhibited the uptake of LPS, and LPS also inhibited the uptake of CnB. These results indicate that the uptake of exogenous CnB did not occur through LPS and that CnB was not a chaperone of LPS. Thus, we conclude that TLR4 receptor complexes were required for the recognition and internalization of exogenous CnB. CnB could be a potential endogenous ligand of TLR4 and function as an agonist of TLR4. These properties of CnB support its potential for development as an anti-cancer drug.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


Similar articles
-
Sialyl residues modulate LPS-mediated signaling through the Toll-like receptor 4 complex.PLoS One. 2012;7(4):e32359. doi: 10.1371/journal.pone.0032359. Epub 2012 Apr 9. PLoS One. 2012. PMID: 22496731 Free PMC article.
-
Dynamic lipopolysaccharide transfer cascade to TLR4/MD2 complex via LBP and CD14.BMB Rep. 2017 Feb;50(2):55-57. doi: 10.5483/bmbrep.2017.50.2.011. BMB Rep. 2017. PMID: 28115037 Free PMC article.
-
A Single Step in vitro Bioassay Mimicking TLR4-LPS Pathway and the Role of MD2 and CD14 Coreceptors.Front Immunol. 2020 Jan 24;11:5. doi: 10.3389/fimmu.2020.00005. eCollection 2020. Front Immunol. 2020. PMID: 32038655 Free PMC article.
-
Quantitative single-molecule imaging of TLR4 reveals ligand-specific receptor dimerization.Sci Signal. 2017 Oct 31;10(503):eaan1308. doi: 10.1126/scisignal.aan1308. Sci Signal. 2017. PMID: 29089449
-
Interaction between polysaccharides and toll-like receptor 4: Primary structural role, immune balance perspective, and 3D interaction model hypothesis.Food Chem. 2022 Apr 16;374:131586. doi: 10.1016/j.foodchem.2021.131586. Epub 2021 Nov 11. Food Chem. 2022. PMID: 34839969 Review.
Cited by
-
In Silico Characterization of Calcineurin from Pathogenic Obligate Intracellular Trypanosomatids: Potential New Biological Roles.Biomolecules. 2021 Sep 7;11(9):1322. doi: 10.3390/biom11091322. Biomolecules. 2021. PMID: 34572535 Free PMC article.
-
rhCNB Improves Cyclophosphamide-Induced Immunodeficiency in BALB/c Mice.Evid Based Complement Alternat Med. 2022 Sep 27;2022:4891399. doi: 10.1155/2022/4891399. eCollection 2022. Evid Based Complement Alternat Med. 2022. PMID: 36204132 Free PMC article.
-
The genetically engineered drug rhCNB induces apoptosis via a mitochondrial route in tumor cells.Oncotarget. 2017 Jul 22;8(39):65876-65888. doi: 10.18632/oncotarget.19507. eCollection 2017 Sep 12. Oncotarget. 2017. PMID: 29029479 Free PMC article.
-
β-chitosan attenuates hepatic macrophage-driven inflammation and reverses aging-related cognitive impairment.iScience. 2024 Aug 20;27(9):110766. doi: 10.1016/j.isci.2024.110766. eCollection 2024 Sep 20. iScience. 2024. PMID: 39280626 Free PMC article.
-
Pyroptosis in renal inflammation and fibrosis: current knowledge and clinical significance.Cell Death Dis. 2023 Jul 27;14(7):472. doi: 10.1038/s41419-023-06005-6. Cell Death Dis. 2023. PMID: 37500614 Free PMC article. Review.
References
-
- Hemenway C. S. & Heitman J. Calcineurin. Structure, function, and inhibition. Cell Biochem Biophys. 30, 115–151 (1999). - PubMed
-
- Li W. & Handschumacher R. E. Identification of two calcineurin B-binding proteins: tubulin and heat shock protein 60. Biochim Biophys Acta. 1599, 72–81 (2002). - PubMed
-
- Saeki M. et al.. Calcineurin potentiates the activation of procaspase-3 by accelerating its proteolytic maturation. J Biol Chem. 282, 11786–11794 (2007). - PubMed
-
- Li N., Zhang Z., Zhang W. & Wei Q. Calcineurin B subunit interacts with proteasome subunit alpha type 7 and represses hypoxia-inducible factor-1α activity via the proteasome pathway. Biochem Biophys Res Commun. 405, 468–472 (2011). - PubMed
-
- Jing L. et al.. Calcineurin subunit B activates dendritic cells and acts as a cancer vaccine adjuvant. Int Immunol. 23, 327–334 (2011). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

