Divergence of RNA polymerase α subunits in angiosperm plastid genomes is mediated by genomic rearrangement
- PMID: 27087667
- PMCID: PMC4834550
- DOI: 10.1038/srep24595
Divergence of RNA polymerase α subunits in angiosperm plastid genomes is mediated by genomic rearrangement
Abstract
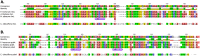
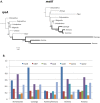
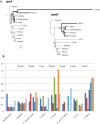
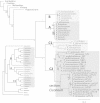
Genes for the plastid-encoded RNA polymerase (PEP) persist in the plastid genomes of all photosynthetic angiosperms. However, three unrelated lineages (Annonaceae, Passifloraceae and Geraniaceae) have been identified with unusually divergent open reading frames (ORFs) in the conserved region of rpoA, the gene encoding the PEP α subunit. We used sequence-based approaches to evaluate whether these genes retain function. Both gene sequences and complete plastid genome sequences were assembled and analyzed from each of the three angiosperm families. Multiple lines of evidence indicated that the rpoA sequences are likely functional despite retaining as low as 30% nucleotide sequence identity with rpoA genes from outgroups in the same angiosperm order. The ratio of non-synonymous to synonymous substitutions indicated that these genes are under purifying selection, and bioinformatic prediction of conserved domains indicated that functional domains are preserved. One of the lineages (Pelargonium, Geraniaceae) contains species with multiple rpoA-like ORFs that show evidence of ongoing inter-paralog gene conversion. The plastid genomes containing these divergent rpoA genes have experienced extensive structural rearrangement, including large expansions of the inverted repeat. We propose that illegitimate recombination, not positive selection, has driven the divergence of rpoA.
Figures
Similar articles
-
Highly accelerated rates of genomic rearrangements and nucleotide substitutions in plastid genomes of Passiflora subgenus Decaloba.Mol Phylogenet Evol. 2019 Sep;138:53-64. doi: 10.1016/j.ympev.2019.05.030. Epub 2019 May 23. Mol Phylogenet Evol. 2019. PMID: 31129347
-
Transcription of plastid genes is modulated by two nuclear-encoded alpha subunits of plastid RNA polymerase in the moss Physcomitrella patens.Plant J. 2007 Nov;52(4):730-41. doi: 10.1111/j.1365-313X.2007.03270.x. Epub 2007 Sep 25. Plant J. 2007. PMID: 17894784
-
Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: rearrangements, repeats, and codon usage.Mol Biol Evol. 2011 Jan;28(1):583-600. doi: 10.1093/molbev/msq229. Epub 2010 Aug 30. Mol Biol Evol. 2011. PMID: 20805190
-
Chloroplast RNA polymerases: Role in chloroplast biogenesis.Biochim Biophys Acta. 2015 Sep;1847(9):761-9. doi: 10.1016/j.bbabio.2015.02.004. Epub 2015 Feb 11. Biochim Biophys Acta. 2015. PMID: 25680513 Review.
-
[Plastid RNA polymerases].Mol Biol (Mosk). 2005 Sep-Oct;39(5):762-75. Mol Biol (Mosk). 2005. PMID: 16240710 Review. Russian.
Cited by
-
Lineage-Specific Variation in IR Boundary Shift Events, Inversions, and Substitution Rates among Caprifoliaceae s.l. (Dipsacales) Plastomes.Int J Mol Sci. 2021 Sep 28;22(19):10485. doi: 10.3390/ijms221910485. Int J Mol Sci. 2021. PMID: 34638831 Free PMC article.
-
Complete chloroplast genomes and comparative analysis of Ligustrum species.Sci Rep. 2023 Jan 5;13(1):212. doi: 10.1038/s41598-022-26884-7. Sci Rep. 2023. PMID: 36604557 Free PMC article.
-
Insights into Comparative Genomics, Codon Usage Bias, and Phylogenetic Relationship of Species from Biebersteiniaceae and Nitrariaceae Based on Complete Chloroplast Genomes.Plants (Basel). 2020 Nov 18;9(11):1605. doi: 10.3390/plants9111605. Plants (Basel). 2020. PMID: 33218207 Free PMC article.
-
Interspecific Hybrids Between Pelargonium × hortorum and Species From P. Section Ciconium Reveal Biparental Plastid Inheritance and Multi-Locus Cyto-Nuclear Incompatibility.Front Plant Sci. 2020 Dec 18;11:614871. doi: 10.3389/fpls.2020.614871. eCollection 2020. Front Plant Sci. 2020. PMID: 33391328 Free PMC article.
-
Evolutionary dynamics of the chloroplast genome sequences of six Colobanthus species.Sci Rep. 2020 Jul 13;10(1):11522. doi: 10.1038/s41598-020-68563-5. Sci Rep. 2020. PMID: 32661280 Free PMC article.
References
-
- Downie S. R. & Palmer J. D. in Molecular Systematics of Plants (eds Soltis P. S., Soltis D. E. & Doyle J. J.) Ch. 2, 14–35 (Springer, 1992).
-
- Doyle J. J., Doyle J. L. & Palmer J. D. Multiple independent losses of two genes and one intron from legume chloroplast genomes. Syst Bot. 20, 272–294 (1995).
-
- Bock R. In Cell and Molecular Biology of Plastids (ed Bock R.) Vol. 19, 29–63 (Springer, 2007).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

