Ciliate Paramecium is a natural reservoir of Legionella pneumophila
- PMID: 27079173
- PMCID: PMC4832178
- DOI: 10.1038/srep24322
Ciliate Paramecium is a natural reservoir of Legionella pneumophila
Abstract
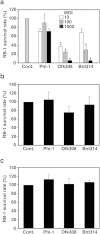
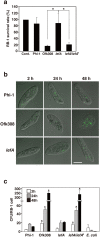
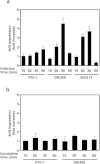
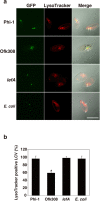
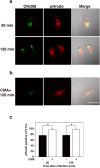
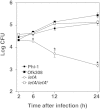
Legionella pneumophila, the causative agent of Legionnaires' disease, replicates within alveolar macrophages and free-living amoebae. However, the lifestyle of L. pneumophila in the environment remains largely unknown. Here we established a novel natural host model of L. pneumophila endosymbiosis using the ciliate Paramecium caudatum. We also identified Legionella endosymbiosis-modulating factor A (LefA), which contributes to the change in life stage from endosymbiosis to host lysis, enabling escape to the environment. We isolated L. pneumophila strains from the environment, and they exhibited cytotoxicity toward P. caudatum and induced host lysis. Acidification of the Legionella-containing vacuole (LCV) was inhibited, and enlarged LCVs including numerous bacteria were observed in P. caudatum infected with L. pneumophila. An isogenic L. pneumophila lefA mutant exhibited decreased cytotoxicity toward P. caudatum and impaired the modification of LCVs, resulting in the establishment of endosymbiosis between them. Our results suggest that L. pneumophila may have a mechanism to switch their endosymbiosis in protistan hosts in the environment.
Figures
Similar articles
-
From Many Hosts, One Accidental Pathogen: The Diverse Protozoan Hosts of Legionella.Front Cell Infect Microbiol. 2017 Nov 30;7:477. doi: 10.3389/fcimb.2017.00477. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 29250488 Free PMC article. Review.
-
Identification of novel Legionella genes required for endosymbiosis in Paramecium based on comparative genome analysis with Holospora spp.FEMS Microbiol Ecol. 2018 Nov 1;94(11). doi: 10.1093/femsec/fiy162. FEMS Microbiol Ecol. 2018. PMID: 30124811
-
Activation of Ran GTPase by a Legionella effector promotes microtubule polymerization, pathogen vacuole motility and infection.PLoS Pathog. 2013 Sep;9(9):e1003598. doi: 10.1371/journal.ppat.1003598. Epub 2013 Sep 19. PLoS Pathog. 2013. PMID: 24068924 Free PMC article.
-
InSeq analysis of defined Legionella pneumophila libraries identifies a transporter-encoding gene cluster important for intracellular replication in mammalian hosts.mBio. 2024 Nov 13;15(11):e0195524. doi: 10.1128/mbio.01955-24. Epub 2024 Oct 4. mBio. 2024. PMID: 39365064 Free PMC article.
-
The life stage-specific pathometabolism of Legionella pneumophila.FEBS Lett. 2016 Nov;590(21):3868-3886. doi: 10.1002/1873-3468.12326. Epub 2016 Aug 18. FEBS Lett. 2016. PMID: 27455397 Review.
Cited by
-
Chasing Waterborne Pathogens in Antarctic Human-Made and Natural Environments, with Special Reference to Legionella spp.Appl Environ Microbiol. 2021 Jan 4;87(2):e02247-20. doi: 10.1128/AEM.02247-20. Print 2021 Jan 4. Appl Environ Microbiol. 2021. PMID: 33097517 Free PMC article.
-
The core microbiome of sessile ciliate Stentor coeruleus is not shaped by the environment.Sci Rep. 2019 Aug 6;9(1):11356. doi: 10.1038/s41598-019-47701-8. Sci Rep. 2019. PMID: 31388025 Free PMC article.
-
Stagnation arising through intermittent usage is associated with increased viable but non culturable Legionella and amoeba hosts in a hospital water system.Front Cell Infect Microbiol. 2023 Jun 7;13:1190631. doi: 10.3389/fcimb.2023.1190631. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37351181 Free PMC article.
-
From Many Hosts, One Accidental Pathogen: The Diverse Protozoan Hosts of Legionella.Front Cell Infect Microbiol. 2017 Nov 30;7:477. doi: 10.3389/fcimb.2017.00477. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 29250488 Free PMC article. Review.
-
Protein sociology of ProA, Mip and other secreted virulence factors at the Legionella pneumophila surface.Front Cell Infect Microbiol. 2023 Mar 2;13:1140688. doi: 10.3389/fcimb.2023.1140688. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 36936764 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

