Characterization of Lgr5+ progenitor cell transcriptomes in the apical and basal turns of the mouse cochlea
- PMID: 27070092
- PMCID: PMC5173047
- DOI: 10.18632/oncotarget.8636
Characterization of Lgr5+ progenitor cell transcriptomes in the apical and basal turns of the mouse cochlea
Abstract
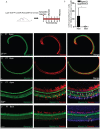
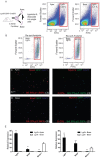
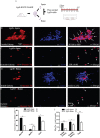
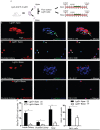
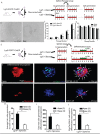
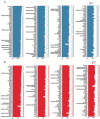
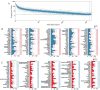
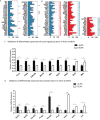
Lgr5+ supporting cells (SCs) are enriched hair cell (HC) progenitors in the cochlea, and several studies have shown a difference in the proliferation and HC regeneration ability of SCs between the apical and basal turns. However, the detailed differences between the transcriptomes of the apical and basal Lgr5+ SCs have not yet been investigated. We found that when isolated by FACS, Lgr5+ cells from the apex generated significantly more HCs and had significantly higher proliferation and mitotic HC regeneration ability compared to those from the base. Next, we used microarray analysis to determine the transcriptome expression profiles of Lgr5+ progenitors from the apex and the base. We first analyzed the genes that were enriched and differentially expressed in Lgr5+ progenitors from the apex and the base. Then we analyzed the cell cycle genes and the transcription factors that might regulate the proliferation and differentiation of Lgr5+ progenitors. Lastly, to further analyze the role of differentially expressed genes and to gain an overall view of the gene network in cochlear HC regeneration, we created a protein-protein interaction network. Our datasets suggest the possible genes that might regulate the proliferation and HC regeneration ability of Lgr5+ progenitors, and these genes might provide new therapeutic targets for HC regeneration in the future.
Keywords: Lgr5; Wnt signaling; cochlea; proliferation; regeneration.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
Characterization of the Transcriptomes of Lgr5+ Hair Cell Progenitors and Lgr5- Supporting Cells in the Mouse Cochlea.Front Mol Neurosci. 2017 Apr 26;10:122. doi: 10.3389/fnmol.2017.00122. eCollection 2017. Front Mol Neurosci. 2017. PMID: 28491023 Free PMC article.
-
Characterization of Lgr5+ Progenitor Cell Transcriptomes after Neomycin Injury in the Neonatal Mouse Cochlea.Front Mol Neurosci. 2017 Jul 4;10:213. doi: 10.3389/fnmol.2017.00213. eCollection 2017. Front Mol Neurosci. 2017. PMID: 28725177 Free PMC article.
-
Wnt activation followed by Notch inhibition promotes mitotic hair cell regeneration in the postnatal mouse cochlea.Oncotarget. 2016 Oct 11;7(41):66754-66768. doi: 10.18632/oncotarget.11479. Oncotarget. 2016. PMID: 27564256 Free PMC article.
-
Postnatal development, maturation and aging in the mouse cochlea and their effects on hair cell regeneration.Hear Res. 2013 Mar;297:68-83. doi: 10.1016/j.heares.2012.11.009. Epub 2012 Nov 16. Hear Res. 2013. PMID: 23164734 Free PMC article. Review.
-
Hair cell fate decisions in cochlear development and regeneration.Hear Res. 2010 Jul;266(1-2):18-25. doi: 10.1016/j.heares.2010.04.012. Epub 2010 May 5. Hear Res. 2010. PMID: 20438823 Free PMC article. Review.
Cited by
-
FGF22 deletion causes hidden hearing loss by affecting the function of inner hair cell ribbon synapses.Front Mol Neurosci. 2022 Jul 28;15:922665. doi: 10.3389/fnmol.2022.922665. eCollection 2022. Front Mol Neurosci. 2022. PMID: 35966010 Free PMC article.
-
LGR4 and LGR5 Regulate Hair Cell Differentiation in the Sensory Epithelium of the Developing Mouse Cochlea.Front Cell Neurosci. 2016 Aug 5;10:186. doi: 10.3389/fncel.2016.00186. eCollection 2016. Front Cell Neurosci. 2016. PMID: 27559308 Free PMC article.
-
Frizzled-9+ Supporting Cells Are Progenitors for the Generation of Hair Cells in the Postnatal Mouse Cochlea.Front Mol Neurosci. 2019 Jul 31;12:184. doi: 10.3389/fnmol.2019.00184. eCollection 2019. Front Mol Neurosci. 2019. PMID: 31427926 Free PMC article.
-
MECOM promotes supporting cell proliferation and differentiation in cochlea.J Otol. 2022 Apr;17(2):59-66. doi: 10.1016/j.joto.2021.11.002. Epub 2021 Nov 24. J Otol. 2022. PMID: 35949554 Free PMC article.
-
Hedgehog Signaling Promotes the Proliferation and Subsequent Hair Cell Formation of Progenitor Cells in the Neonatal Mouse Cochlea.Front Mol Neurosci. 2017 Dec 21;10:426. doi: 10.3389/fnmol.2017.00426. eCollection 2017. Front Mol Neurosci. 2017. PMID: 29311816 Free PMC article.
References
-
- Corwin JT, Cotanche DA. Regeneration of sensory hair cells after acoustic trauma. Science. 1988;240:1772–1774. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

