5-Hydroxymethylation-associated epigenetic modifiers of Alzheimer's disease modulate Tau-induced neurotoxicity
- PMID: 27060332
- PMCID: PMC5181627
- DOI: 10.1093/hmg/ddw109
5-Hydroxymethylation-associated epigenetic modifiers of Alzheimer's disease modulate Tau-induced neurotoxicity
Abstract
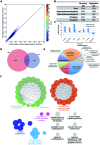
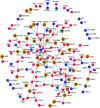
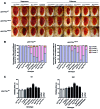
Alzheimer's disease (AD) is a chronic neurodegenerative disorder characterized by progressive deterioration of cognitive function. Pathogenesis of AD is incompletely understood; evidence suggests a role for epigenetic regulation, in particular the cytosine modifications 5-methylcytosine and 5-hydroxymethylcytosine (5hmC). 5hmC is enriched in the nervous system and displays neurodevelopment and age-related changes. To determine the role of 5hmC in AD, we performed genome-wide analyses of 5hmC in DNA from prefrontal cortex of post-mortem AD patients, and RNA-Seq to correlate changes in 5hmC with transcriptional changes. We identified 325 genes containing differentially hydroxymethylated loci (DhMLs) in both discovery and replication datasets. These are enriched for pathways involved in neuron projection development and neurogenesis. Of these, 140 showed changes in gene expression. Proteins encoded by these genes form direct protein-protein interactions with AD-associated genes, expanding the network of genes implicated in AD. We identified AD-associated single nucleotide polymorphisms (SNPs) located within or near DhMLs, suggesting these SNPs may identify regions of epigenetic gene regulation that play a role in AD pathogenesis. Finally, using an existing AD fly model, we showed some of these genes modulate AD-associated toxicity. Our data implicate neuronal projection development and neurogenesis pathways as potential targets in AD. By incorporating epigenomic and transcriptomic data with genome-wide association studies data, with verification in the Drosophila model, we can expand the known network of genes involved in disease pathogenesis and identify epigenetic modifiers of Alzheimer's disease.
© The Author 2016. Published by Oxford University Press. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures


Similar articles
-
Global changes in DNA methylation and hydroxymethylation in Alzheimer's disease human brain.Neurobiol Aging. 2014 Jun;35(6):1334-44. doi: 10.1016/j.neurobiolaging.2013.11.031. Epub 2013 Dec 4. Neurobiol Aging. 2014. PMID: 24387984
-
Genome-wide alteration of 5-hydroxymenthylcytosine in a mouse model of Alzheimer's disease.BMC Genomics. 2016 May 20;17:381. doi: 10.1186/s12864-016-2731-1. BMC Genomics. 2016. PMID: 27207465 Free PMC article.
-
5-Hydroxymethylcytosine Signatures in Circulating Cell-Free DNA as Diagnostic Biomarkers for Late-Onset Alzheimer's Disease.J Alzheimers Dis. 2022;85(2):573-585. doi: 10.3233/JAD-215217. J Alzheimers Dis. 2022. PMID: 34864677
-
DNA Modifications and Alzheimer's Disease.Adv Exp Med Biol. 2017;978:303-319. doi: 10.1007/978-3-319-53889-1_16. Adv Exp Med Biol. 2017. PMID: 28523553 Review.
-
Epigenetics in Alzheimer's disease: a focus on DNA modifications.Curr Pharm Des. 2011;17(31):3398-412. doi: 10.2174/138161211798072544. Curr Pharm Des. 2011. PMID: 21902668 Review.
Cited by
-
Epigenetic Regulations in Neural Stem Cells and Neurological Diseases.Stem Cells Int. 2018 Mar 18;2018:6087143. doi: 10.1155/2018/6087143. eCollection 2018. Stem Cells Int. 2018. PMID: 29743892 Free PMC article. Review.
-
DNA methylation alterations in Alzheimer's disease.Environ Epigenet. 2017 Jun 6;3(2):dvx008. doi: 10.1093/eep/dvx008. eCollection 2017 May. Environ Epigenet. 2017. PMID: 29492310 Free PMC article. Review.
-
Transcriptomic Analyses for Identification and Prioritization of Genes Associated With Alzheimer's Disease in Humans.Front Bioeng Biotechnol. 2020 Feb 21;8:31. doi: 10.3389/fbioe.2020.00031. eCollection 2020. Front Bioeng Biotechnol. 2020. PMID: 32154224 Free PMC article.
-
Blood DNA methylation as a potential biomarker of dementia: A systematic review.Alzheimers Dement. 2018 Jan;14(1):81-103. doi: 10.1016/j.jalz.2017.10.002. Epub 2017 Nov 8. Alzheimers Dement. 2018. PMID: 29127806 Free PMC article.
-
Opening up the DNA methylome of dementia.Mol Psychiatry. 2017 Apr;22(4):485-496. doi: 10.1038/mp.2016.242. Epub 2017 Jan 3. Mol Psychiatry. 2017. PMID: 28044062 Free PMC article. Review.
References
-
- Barker W.W., Luis C.A., Kashuba A., Luis M., Harwood D.G., Loewenstein D., Waters C., Jimison P., Shepherd E., Sevush S. et al. (2002) Relative frequencies of Alzheimer disease, Lewy body, vascular and frontotemporal dementia, and Hippocampal sclerosis in the State of Florida Brain Bank. Alzheimer Dis. Assoc. Disord., 16, 203–212. - PubMed
-
- Hardy J., Selkoe D.J. (2002) The amyloid hypothesis of Alzheimer's disease, progress and problems on the road to therapeutics. Science, 297, 353–356. - PubMed
-
- Blennow K., de Leon M.J., Zetterberg H. (2006) Alzheimer's disease. Lancet, 368, 387–403. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

