PolySUMOylation by Siz2 and Mms21 triggers relocation of DNA breaks to nuclear pores through the Slx5/Slx8 STUbL
- PMID: 27056668
- PMCID: PMC4840299
- DOI: 10.1101/gad.277665.116
PolySUMOylation by Siz2 and Mms21 triggers relocation of DNA breaks to nuclear pores through the Slx5/Slx8 STUbL
Abstract
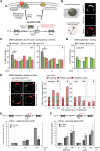
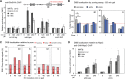
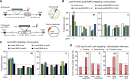
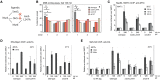
High-resolution imaging shows that persistent DNA damage in budding yeast localizes in distinct perinuclear foci for repair. The signals that trigger DNA double-strand break (DSB) relocation or determine their destination are unknown. We show here that DSB relocation to the nuclear envelope depends on SUMOylation mediated by the E3 ligases Siz2 and Mms21. In G1, a polySUMOylation signal deposited coordinately by Mms21 and Siz2 recruits the SUMO targeted ubiquitin ligase Slx5/Slx8 to persistent breaks. Both Slx5 and Slx8 are necessary for damage relocation to nuclear pores. When targeted to an undamaged locus, however, Slx5 alone can mediate relocation in G1-phase cells, bypassing the requirement for polySUMOylation. In contrast, in S-phase cells, monoSUMOylation mediated by the Rtt107-stabilized SMC5/6-Mms21 E3 complex drives DSBs to the SUN domain protein Mps3 in a manner independent of Slx5. Slx5/Slx8 and binding to pores favor repair by ectopic break-induced replication and imprecise end-joining.
Keywords: DNA damage; Mms21; SUMO; Siz2; Slx5; nuclear organization; nuclear pores.
© 2016 Horigome et al.; Published by Cold Spring Harbor Laboratory Press.
Figures

Similar articles
-
Disruption of SUMO-targeted ubiquitin ligases Slx5-Slx8/RNF4 alters RecQ-like helicase Sgs1/BLM localization in yeast and human cells.DNA Repair (Amst). 2015 Feb;26:1-14. doi: 10.1016/j.dnarep.2014.12.004. Epub 2014 Dec 26. DNA Repair (Amst). 2015. PMID: 25588990 Free PMC article.
-
Regulation of recombination at yeast nuclear pores controls repair and triplet repeat stability.Genes Dev. 2015 May 15;29(10):1006-17. doi: 10.1101/gad.256404.114. Epub 2015 May 4. Genes Dev. 2015. PMID: 25940904 Free PMC article.
-
Activation of the Slx5-Slx8 ubiquitin ligase by poly-small ubiquitin-like modifier conjugates.J Biol Chem. 2008 Jul 18;283(29):19912-21. doi: 10.1074/jbc.M802690200. Epub 2008 May 22. J Biol Chem. 2008. PMID: 18499666 Free PMC article.
-
Nuclear organization in genome stability: SUMO connections.Cell Res. 2011 Mar;21(3):474-85. doi: 10.1038/cr.2011.31. Epub 2011 Feb 15. Cell Res. 2011. PMID: 21321608 Free PMC article. Review.
-
SUMO-targeted ubiquitin ligases.Biochim Biophys Acta. 2014 Jan;1843(1):75-85. doi: 10.1016/j.bbamcr.2013.08.022. Epub 2013 Sep 7. Biochim Biophys Acta. 2014. PMID: 24018209 Review.
Cited by
-
The S phase checkpoint promotes the Smc5/6 complex dependent SUMOylation of Pol2, the catalytic subunit of DNA polymerase ε.PLoS Genet. 2019 Nov 25;15(11):e1008427. doi: 10.1371/journal.pgen.1008427. eCollection 2019 Nov. PLoS Genet. 2019. PMID: 31765407 Free PMC article.
-
Location, Location, Location: The Role of Nuclear Positioning in the Repair of Collapsed Forks and Protection of Genome Stability.Genes (Basel). 2020 Jun 9;11(6):635. doi: 10.3390/genes11060635. Genes (Basel). 2020. PMID: 32526925 Free PMC article. Review.
-
The non-homologous end-joining factor Nej1 inhibits resection mediated by Dna2-Sgs1 nuclease-helicase at DNA double strand breaks.J Biol Chem. 2017 Sep 1;292(35):14576-14586. doi: 10.1074/jbc.M117.796011. Epub 2017 Jul 5. J Biol Chem. 2017. PMID: 28679532 Free PMC article.
-
The INO80 remodeller in transcription, replication and repair.Philos Trans R Soc Lond B Biol Sci. 2017 Oct 5;372(1731):20160290. doi: 10.1098/rstb.2016.0290. Philos Trans R Soc Lond B Biol Sci. 2017. PMID: 28847827 Free PMC article. Review.
-
The nuclear pore primes recombination-dependent DNA synthesis at arrested forks by promoting SUMO removal.Nat Commun. 2020 Nov 6;11(1):5643. doi: 10.1038/s41467-020-19516-z. Nat Commun. 2020. PMID: 33159083 Free PMC article.
References
-
- Agmon N, Liefshitz B, Zimmer C, Fabre E, Kupiec M. 2013. Effect of nuclear architecture on the efficiency of double-strand break repair. Nat Cell Biol 15: 694–699. - PubMed
-
- Bennett CB, Lewis LK, Karthikeyan G, Lobachev KS, Jin YH, Sterling JF, Snipe JR, Resnick MA. 2001. Genes required for ionizing radiation resistance in yeast. Nat Genet 29: 426–434. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
