Direct-methods structure determination of a trypanosome RNA-editing substrate fragment with translational pseudosymmetry
- PMID: 27050127
- PMCID: PMC4822560
- DOI: 10.1107/S2059798316001224
Direct-methods structure determination of a trypanosome RNA-editing substrate fragment with translational pseudosymmetry
Abstract
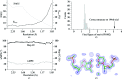
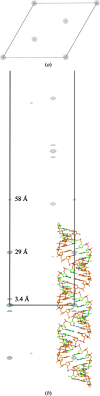
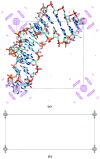
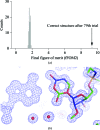
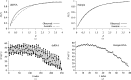
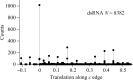
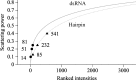
Using direct methods starting from random phases, the crystal structure of a 32-base-pair RNA (675 non-H RNA atoms in the asymmetric unit) was determined using only the native diffraction data (resolution limit 1.05 Å) and the computer program SIR2014. The almost three helical turns of the RNA in the asymmetric unit introduced partial or imperfect translational pseudosymmetry (TPS) that modulated the intensities when averaged by the l Miller indices but still escaped automated detection. Almost six times as many random phase sets had to be tested on average to reach a correct structure compared with a similar-sized RNA hairpin (27 nucleotides, 580 non-H RNA atoms) without TPS. More sensitive methods are needed for the automated detection of partial TPS.
Keywords: Patterson analysis; RNA structure determination; ab initio phasing; helical symmetry; noncrystallographic symmetry.
Figures



Similar articles
-
Fusion RNAs in crystallographic studies of double-stranded RNA from trypanosome RNA editing.Methods Mol Biol. 2015;1240:191-216. doi: 10.1007/978-1-4939-1896-6_14. Methods Mol Biol. 2015. PMID: 25352146
-
The crystal structure of an oligo(U):pre-mRNA duplex from a trypanosome RNA editing substrate.RNA. 2011 Oct;17(10):1870-83. doi: 10.1261/rna.2880311. Epub 2011 Aug 30. RNA. 2011. PMID: 21878548 Free PMC article.
-
Structural studies of a double-stranded RNA from trypanosome RNA editing by small-angle X-ray scattering.Methods Mol Biol. 2015;1240:165-89. doi: 10.1007/978-1-4939-1896-6_13. Methods Mol Biol. 2015. PMID: 25352145
-
Ab initio phase determination and phase extension using non-crystallographic symmetry.Curr Opin Struct Biol. 1995 Oct;5(5):650-5. doi: 10.1016/0959-440x(95)80058-1. Curr Opin Struct Biol. 1995. PMID: 8574701 Review.
-
ACORN: a review.Acta Crystallogr D Biol Crystallogr. 2006 Aug;62(Pt 8):901-8. doi: 10.1107/S0907444906008122. Epub 2006 Jul 18. Acta Crystallogr D Biol Crystallogr. 2006. PMID: 16855307 Review.
Cited by
-
Engineering Crystal Packing in RNA Structures I: Past and Future Strategies for Engineering RNA Packing in Crystals.Crystals (Basel). 2021 Aug;11(8):952. doi: 10.3390/cryst11080952. Epub 2021 Aug 15. Crystals (Basel). 2021. PMID: 34745656 Free PMC article.
-
Genetic identification of the functional surface for RNA binding by Escherichia coli ProQ.Nucleic Acids Res. 2020 May 7;48(8):4507-4520. doi: 10.1093/nar/gkaa144. Nucleic Acids Res. 2020. PMID: 32170306 Free PMC article.
-
The structure of denisovite, a fibrous nanocrystalline polytypic disordered 'very complex' silicate, studied by a synergistic multi-disciplinary approach employing methods of electron crystallography and X-ray powder diffraction.IUCrJ. 2017 Mar 8;4(Pt 3):223-242. doi: 10.1107/S2052252517002585. eCollection 2017 May 1. IUCrJ. 2017. PMID: 28512570 Free PMC article.
-
Targeting RNA Structure to Inhibit Editing in Trypanosomes.Int J Mol Sci. 2023 Jun 14;24(12):10110. doi: 10.3390/ijms241210110. Int J Mol Sci. 2023. PMID: 37373258 Free PMC article.
References
-
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221. - PubMed
-
- Böhme, R. (1982). Acta Cryst. A38, 318–326.
-
- Burla, M. C., Caliandro, R., Carrozzini, B., Cascarano, G. L., Cuocci, C., Giacovazzo, C., Mallamo, M., Mazzone, A. & Polidori, G. (2015). J. Appl. Cryst. 48, 306–309.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

