Giant Viruses of Amoebas: An Update
- PMID: 27047465
- PMCID: PMC4801854
- DOI: 10.3389/fmicb.2016.00349
Giant Viruses of Amoebas: An Update
Abstract
During the 12 past years, five new or putative virus families encompassing several members, namely Mimiviridae, Marseilleviridae, pandoraviruses, faustoviruses, and virophages were described. In addition, Pithovirus sibericum and Mollivirus sibericum represent type strains of putative new giant virus families. All these viruses were isolated using amoebal coculture methods. These giant viruses were linked by phylogenomic analyses to other large DNA viruses. They were then proposed to be classified in a new viral order, the Megavirales, on the basis of their common origin, as shown by a set of ancestral genes encoding key viral functions, a common virion architecture, and shared major biological features including replication inside cytoplasmic factories. Megavirales is increasingly demonstrated to stand in the tree of life aside Bacteria, Archaea, and Eukarya, and the megavirus ancestor is suspected to be as ancient as cellular ancestors. In addition, giant amoebal viruses are visible under a light microscope and display many phenotypic and genomic features not found in other viruses, while they share other characteristics with parasitic microbes. Moreover, these organisms appear to be common inhabitants of our biosphere, and mimiviruses and marseilleviruses were isolated from human samples and associated to diseases. In the present review, we describe the main features and recent findings on these giant amoebal viruses and virophages.
Keywords: 4th TRUC; Acanthamoeba; Megavirales; amoeba; giant virus; mimivirus; virophage.
Figures


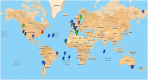
 , indicates location of samples from which an amoebal virus was isolated;
, indicates location of samples from which an amoebal virus was isolated;  , indicates location of samples from which reads related to an amoebal virus were generated by metagenomics;
, indicates location of samples from which reads related to an amoebal virus were generated by metagenomics;  , indicates the discovery of giant viral particles for which a virus could not be isolated. Blue color indicates environmental samples; green color indicates human samples; red color indicates animal (non-human) samples. This figure is a screenshot of a goggle map that is freely available at the following URL:
, indicates the discovery of giant viral particles for which a virus could not be isolated. Blue color indicates environmental samples; green color indicates human samples; red color indicates animal (non-human) samples. This figure is a screenshot of a goggle map that is freely available at the following URL: Similar articles
-
Pithovirus sibericum, a new bona fide member of the "Fourth TRUC" club.Front Microbiol. 2015 Aug 4;6:722. doi: 10.3389/fmicb.2015.00722. eCollection 2015. Front Microbiol. 2015. PMID: 26300849 Free PMC article.
-
Mimivirus: leading the way in the discovery of giant viruses of amoebae.Nat Rev Microbiol. 2017 Apr;15(4):243-254. doi: 10.1038/nrmicro.2016.197. Epub 2017 Feb 27. Nat Rev Microbiol. 2017. PMID: 28239153 Free PMC article. Review.
-
A New Zamilon-like Virophage Partial Genome Assembled from a Bioreactor Metagenome.Front Microbiol. 2015 Nov 27;6:1308. doi: 10.3389/fmicb.2015.01308. eCollection 2015. Front Microbiol. 2015. PMID: 26640459 Free PMC article.
-
Characterization of Mollivirus kamchatka, the First Modern Representative of the Proposed Molliviridae Family of Giant Viruses.J Virol. 2020 Mar 31;94(8):e01997-19. doi: 10.1128/JVI.01997-19. Print 2020 Mar 31. J Virol. 2020. PMID: 31996429 Free PMC article.
-
Amoebae, Giant Viruses, and Virophages Make Up a Complex, Multilayered Threesome.Front Cell Infect Microbiol. 2018 Jan 11;7:527. doi: 10.3389/fcimb.2017.00527. eCollection 2017. Front Cell Infect Microbiol. 2018. PMID: 29376032 Free PMC article. Review.
Cited by
-
Virome Diversity among Mosquito Populations in a Sub-Urban Region of Marseille, France.Viruses. 2021 Apr 27;13(5):768. doi: 10.3390/v13050768. Viruses. 2021. PMID: 33925487 Free PMC article.
-
New Isolates of Pandoraviruses: Contribution to the Study of Replication Cycle Steps.J Virol. 2019 Feb 19;93(5):e01942-18. doi: 10.1128/JVI.01942-18. Print 2019 Mar 1. J Virol. 2019. PMID: 30541841 Free PMC article.
-
Climate change, melting cryosphere and frozen pathogens: Should we worry…?Environ Sustain (Singap). 2021;4(3):489-501. doi: 10.1007/s42398-021-00184-8. Epub 2021 May 31. Environ Sustain (Singap). 2021. PMID: 38624658 Free PMC article. Review.
-
Virophages of Giant Viruses: An Update at Eleven.Viruses. 2019 Aug 8;11(8):733. doi: 10.3390/v11080733. Viruses. 2019. PMID: 31398856 Free PMC article. Review.
-
Lateral Gene Transfer Between Protozoa-Related Giant Viruses of Family Mimiviridae and Chlamydiae.Evol Bioinform Online. 2018 Jul 17;14:1176934318788337. doi: 10.1177/1176934318788337. eCollection 2018. Evol Bioinform Online. 2018. PMID: 30038484 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

