Doa1 targets ubiquitinated substrates for mitochondria-associated degradation
- PMID: 27044889
- PMCID: PMC4828692
- DOI: 10.1083/jcb.201510098
Doa1 targets ubiquitinated substrates for mitochondria-associated degradation
Abstract
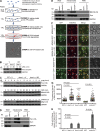
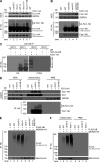
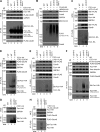
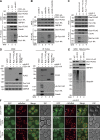
Mitochondria-associated degradation (MAD) mediated by the Cdc48 complex and proteasome degrades ubiquitinated mitochondrial outer-membrane proteins. MAD is critical for mitochondrial proteostasis, but it remains poorly characterized. We identified several mitochondrial Cdc48 substrates and developed a genetic screen assay to uncover regulators of the Cdc48-dependent MAD pathway. Surprisingly, we identified Doa1, a substrate-processing factor of Cdc48 that inhibits the degradation of some Cdc48 substrates, as a critical mediator of the turnover of mitochondrial Cdc48 substrates. Deletion ofDOA1causes the accumulation and mislocalization of substrates on mitochondria. Profiling of Cdc48 cofactors shows that Doa1 and Cdc48(-Ufd1-Npl4)form a functional complex mediating MAD. Biochemically, Doa1 interacts with ubiquitinated substrates and facilitates substrate recruitment to the Cdc48(-Ufd1-Npl4)complex. Functionally, Doa1 is critical for cell survival under mitochondrial oxidative stress, but not ER stress, conditions. Collectively, our results demonstrate the essential role of the Doa1-Cdc48(-Ufd1-Npl4)complex in mitochondrial proteostasis and suggest that Doa1 plays dual roles on the Cdc48 complex.
© 2016 Wu et al.
Figures



Comment in
-
Doa1 is a MAD adaptor for Cdc48.J Cell Biol. 2016 Apr 11;213(1):7-9. doi: 10.1083/jcb.201603078. Epub 2016 Apr 4. J Cell Biol. 2016. PMID: 27044894 Free PMC article.
Similar articles
-
Doa1 is a MAD adaptor for Cdc48.J Cell Biol. 2016 Apr 11;213(1):7-9. doi: 10.1083/jcb.201603078. Epub 2016 Apr 4. J Cell Biol. 2016. PMID: 27044894 Free PMC article.
-
Doa1 is a Cdc48 adapter that possesses a novel ubiquitin binding domain.Mol Cell Biol. 2006 Feb;26(3):822-30. doi: 10.1128/MCB.26.3.822-830.2006. Mol Cell Biol. 2006. PMID: 16428438 Free PMC article.
-
Mitochondrial Surveillance by Cdc48/p97: MAD vs. Membrane Fusion.Int J Mol Sci. 2020 Sep 18;21(18):6841. doi: 10.3390/ijms21186841. Int J Mol Sci. 2020. PMID: 32961852 Free PMC article. Review.
-
A Cdc48 "Retrochaperone" Function Is Required for the Solubility of Retrotranslocated, Integral Membrane Endoplasmic Reticulum-associated Degradation (ERAD-M) Substrates.J Biol Chem. 2017 Feb 24;292(8):3112-3128. doi: 10.1074/jbc.M116.770610. Epub 2017 Jan 11. J Biol Chem. 2017. PMID: 28077573 Free PMC article.
-
Mitochondrial quality control by the ubiquitin-proteasome system.Biochem Soc Trans. 2011 Oct;39(5):1509-13. doi: 10.1042/BST0391509. Biochem Soc Trans. 2011. PMID: 21936843 Review.
Cited by
-
Quality control of the mitochondrial proteome.Nat Rev Mol Cell Biol. 2021 Jan;22(1):54-70. doi: 10.1038/s41580-020-00300-2. Epub 2020 Oct 22. Nat Rev Mol Cell Biol. 2021. PMID: 33093673 Review.
-
Molecular Determinants of Mitochondrial Shape and Function and Their Role in Glaucoma.Antioxid Redox Signal. 2023 May;38(13-15):896-919. doi: 10.1089/ars.2022.0124. Epub 2023 Jan 5. Antioxid Redox Signal. 2023. PMID: 36301938 Free PMC article. Review.
-
OPENER Is a Nuclear Envelope and Mitochondria Localized Protein Required for Cell Cycle Progression in Arabidopsis.Plant Cell. 2019 Jul;31(7):1446-1465. doi: 10.1105/tpc.19.00033. Epub 2019 Apr 25. Plant Cell. 2019. PMID: 31023726 Free PMC article.
-
Mitochondrial inner-membrane protease Yme1 degrades outer-membrane proteins Tom22 and Om45.J Cell Biol. 2018 Jan 2;217(1):139-149. doi: 10.1083/jcb.201702125. Epub 2017 Nov 14. J Cell Biol. 2018. PMID: 29138251 Free PMC article.
-
DLK-1, SEK-3 and PMK-3 Are Required for the Life Extension Induced by Mitochondrial Bioenergetic Disruption in C. elegans.PLoS Genet. 2016 Jul 15;12(7):e1006133. doi: 10.1371/journal.pgen.1006133. eCollection 2016 Jul. PLoS Genet. 2016. PMID: 27420916 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

