Ultraconserved region-containing Transformer 2β4 controls senescence of colon cancer cells
- PMID: 27043659
- PMCID: PMC4848834
- DOI: 10.1038/oncsis.2016.18
Ultraconserved region-containing Transformer 2β4 controls senescence of colon cancer cells
Abstract
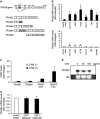
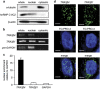
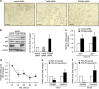
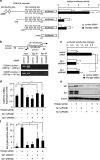
Ultraconserved regions (UCRs) are >200 bp genomic segments with perfect human-to-rodent sequence identity. Transcribed UCRs constitute a new category of noncoding RNAs whose functions remain poorly understood. The human transformer 2β (TRA2B) gene contains a 419-bp UCR spanning the 276-bp exon 2 and its neighboring introns. TRA2B exon 2 has premature stop codons, whereas an exon 2-containing splice variant (TRA2β4) was expressed preferentially in the nuclei of human colon cancer cells. TRA2β4 knockdown p53-independently stimulated CDKN1A transcription and increased p21, resulting in the appearance of senescent cells. Biotin pull-down and RNA immunoprecipitation assays revealed that TRA2β4 interacted with Sp1 through a Sp1-binding sequence (485-GGGG-488) in a stem-loop structure of exon 2. Mutation of this sequence (485-AAGG-488) disrupted the stem-loop structure, blocked the interaction with Sp1 and increased CDKN1A transcription. Overexpression of TRA2β4 significantly decreased CDKN1A mRNA levels and accelerated cell growth, but the introduction of the mutation in the Sp1-binding sequence completely canceled these effects. Taken together, TRA2β4 may sequester Sp1 from occupying promoters of target genes including CDKN1A, promoting cell growth by interrupting the senescence-related gene expression program. This novel function of TRA2β4 may uncover an oncogenic function of transcribed UCRs.
Figures


Similar articles
-
Overexpression of the transcribed ultraconserved region Uc.138 accelerates colon cancer progression.Sci Rep. 2021 Apr 21;11(1):8667. doi: 10.1038/s41598-021-88123-9. Sci Rep. 2021. PMID: 33883665 Free PMC article.
-
Nucleolin facilitates nuclear retention of an ultraconserved region containing TRA2β4 and accelerates colon cancer cell growth.Oncotarget. 2018 Jun 1;9(42):26817-26833. doi: 10.18632/oncotarget.25510. eCollection 2018 Jun 1. Oncotarget. 2018. PMID: 29928487 Free PMC article.
-
HnRNPA1 interacts with G-quadruplex in the TRA2B promoter and stimulates its transcription in human colon cancer cells.Sci Rep. 2019 Jul 16;9(1):10276. doi: 10.1038/s41598-019-46659-x. Sci Rep. 2019. PMID: 31311954 Free PMC article.
-
Highlighting transcribed ultraconserved regions in human diseases.Wiley Interdiscip Rev RNA. 2020 Mar;11(2):e1567. doi: 10.1002/wrna.1567. Epub 2019 Sep 5. Wiley Interdiscip Rev RNA. 2020. PMID: 31489780 Review.
-
Role of transcribed ultraconserved regions in gastric cancer and therapeutic perspectives.World J Gastroenterol. 2022 Jul 7;28(25):2900-2909. doi: 10.3748/wjg.v28.i25.2900. World J Gastroenterol. 2022. PMID: 35978878 Free PMC article. Review.
Cited by
-
Overexpression of the transcribed ultraconserved region Uc.138 accelerates colon cancer progression.Sci Rep. 2021 Apr 21;11(1):8667. doi: 10.1038/s41598-021-88123-9. Sci Rep. 2021. PMID: 33883665 Free PMC article.
-
SPDL1 Is an Independent Predictor of Patient Outcome in Colorectal Cancer.Int J Mol Sci. 2022 Feb 5;23(3):1819. doi: 10.3390/ijms23031819. Int J Mol Sci. 2022. PMID: 35163739 Free PMC article.
-
Expression of TRA2B in endometrial carcinoma and its regulatory roles in endometrial carcinoma cells.Oncol Lett. 2019 Sep;18(3):2455-2463. doi: 10.3892/ol.2019.10553. Epub 2019 Jul 3. Oncol Lett. 2019. PMID: 31452736 Free PMC article.
-
Nucleolin facilitates nuclear retention of an ultraconserved region containing TRA2β4 and accelerates colon cancer cell growth.Oncotarget. 2018 Jun 1;9(42):26817-26833. doi: 10.18632/oncotarget.25510. eCollection 2018 Jun 1. Oncotarget. 2018. PMID: 29928487 Free PMC article.
-
RNA Binding Proteins and Genome Integrity.Int J Mol Sci. 2017 Jun 23;18(7):1341. doi: 10.3390/ijms18071341. Int J Mol Sci. 2017. PMID: 28644387 Free PMC article. Review.
References
-
- Modrek B, Lee C. A genomic view of alternative splicing. Nat Genet 2002; 30: 13–19. - PubMed
-
- Srebrow A, Kornblihtt AR. The connection between splicing and cancer. J Cell Sci 2006; 119: 2635–2641. - PubMed
-
- Venables JP, Klinck R, Koh C, Gervais-Bird J, Bramard A, Inkel L et al. Cancer-associated regulation of alternative splicing. Nat Struct Mol Biol 2009; 16: 670–676. - PubMed
-
- Kondo S, Yamamoto N, Murakami T, Okumura M, Mayeda A, Imaizumi K. Tra2 beta, SF2/ASF and SRp30c modulate the function of an exonic splicing enhancer in exon 10 of tau pre-mRNA. Genes Cells 2004; 9: 121–130. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

