Contributions of DNA repair and damage response pathways to the non-linear genotoxic responses of alkylating agents
- PMID: 27036068
- PMCID: PMC4818947
- DOI: 10.1016/j.mrrev.2015.11.001
Contributions of DNA repair and damage response pathways to the non-linear genotoxic responses of alkylating agents
Abstract
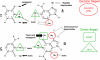
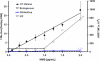
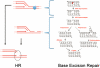
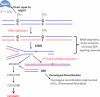
From a risk assessment perspective, DNA-reactive agents are conventionally assumed to have genotoxic risks at all exposure levels, thus applying a linear extrapolation for low-dose responses. New approaches discussed here, including more diverse and sensitive methods for assessing DNA damage and DNA repair, strongly support the existence of measurable regions where genotoxic responses with increasing doses are insignificant relative to control. Model monofunctional alkylating agents have in vitro and in vivo datasets amenable to determination of points of departure (PoDs) for genotoxic effects. A session at the 2013 Society of Toxicology meeting provided an opportunity to survey the progress in understanding the biological basis of empirically-observed PoDs for DNA alkylating agents. Together with the literature published since, this review discusses cellular pathways activated by endogenous and exogenous alkylation DNA damage. Cells have evolved conserved processes that monitor and counteract a spontaneous steady-state level of DNA damage. The ubiquitous network of DNA repair pathways serves as the first line of defense for clearing of the DNA damage and preventing mutation. Other biological pathways discussed here that are activated by genotoxic stress include post-translational activation of cell cycle networks and transcriptional networks for apoptosis/cell death. The interactions of various DNA repair and DNA damage response pathways provide biological bases for the observed PoD behaviors seen with genotoxic compounds. Thus, after formation of DNA adducts, the activation of cellular pathways can lead to the avoidance of a mutagenic outcome. The understanding of the cellular mechanisms acting within the low-dose region will serve to better characterize risks from exposures to DNA-reactive agents at environmentally-relevant concentrations.
Keywords: Biological pathways; DNA damage; DNA repair; Dose–response; Low-dose; Points of departure.
Copyright © 2015 Elsevier B.V. All rights reserved.
Figures




Similar articles
-
Do dose response thresholds exist for genotoxic alkylating agents?Mutagenesis. 2005 Nov;20(6):389-98. doi: 10.1093/mutage/gei054. Epub 2005 Aug 31. Mutagenesis. 2005. PMID: 16135536
-
Assessment of mechanisms driving non-linear dose-response relationships in genotoxicity testing.Mutat Res Rev Mutat Res. 2015 Jan-Mar;763:181-201. doi: 10.1016/j.mrrev.2014.11.001. Epub 2014 Nov 11. Mutat Res Rev Mutat Res. 2015. PMID: 25795120 Review.
-
DNA damage and repair in mutagenesis and carcinogenesis: implications of structure-activity relationships for cross-species extrapolation.Mutat Res. 1996 Jun 12;353(1-2):177-218. doi: 10.1016/0027-5107(96)00032-2. Mutat Res. 1996. PMID: 8692191 Review.
-
Genotoxic thresholds, DNA repair, and susceptibility in human populations.Toxicology. 2010 Dec 30;278(3):305-10. doi: 10.1016/j.tox.2009.11.016. Epub 2009 Nov 22. Toxicology. 2010. PMID: 19932733 Review.
-
International Commission for Protection Against Environmental Mutagens and Carcinogens. The subtlety of alkylating agents in reactions with biological macromolecules.Mutat Res. 1994 Feb 1;305(1):13-32. doi: 10.1016/0027-5107(94)90123-6. Mutat Res. 1994. PMID: 7508544 Review.
Cited by
-
Biological Basis for Threshold Responses to Methylating Agents.Chem Res Toxicol. 2020 Sep 21;33(9):2219-2224. doi: 10.1021/acs.chemrestox.0c00052. Epub 2020 May 27. Chem Res Toxicol. 2020. PMID: 32388971 Free PMC article. Review.
-
The DNA Alkylguanine DNA Alkyltransferase-2 (AGT-2) Of Caenorhabditis Elegans Is Involved In Meiosis And Early Development Under Physiological Conditions.Sci Rep. 2019 May 3;9(1):6889. doi: 10.1038/s41598-019-43394-1. Sci Rep. 2019. PMID: 31053748 Free PMC article.
-
DNA Repair Molecular Beacon assay: a platform for real-time functional analysis of cellular DNA repair capacity.Oncotarget. 2018 Aug 3;9(60):31719-31743. doi: 10.18632/oncotarget.25859. eCollection 2018 Aug 3. Oncotarget. 2018. PMID: 30167090 Free PMC article.
-
Polymerases and DNA Repair in Neurons: Implications in Neuronal Survival and Neurodegenerative Diseases.Front Cell Neurosci. 2022 Jun 30;16:852002. doi: 10.3389/fncel.2022.852002. eCollection 2022. Front Cell Neurosci. 2022. PMID: 35846567 Free PMC article. Review.
-
Mass Spectrometric Analysis of the Active Site Tryptic Peptide of Recombinant O6-Methylguanine-DNA Methyltransferase Following Incubation with Human Colorectal DNA Reveals the Presence of an O6-Alkylguanine Adductome.Chem Res Toxicol. 2023 Dec 18;36(12):1921-1929. doi: 10.1021/acs.chemrestox.3c00207. Epub 2023 Nov 20. Chem Res Toxicol. 2023. PMID: 37983188 Free PMC article.
References
-
- NRC . National Research Council Committee on Improving Risk Analysis Approaches Used by the U. S. EPA Science and Decisions: Advancing Risk Assessment. National Academy of Sciences; Washington D.C.: 2009.
-
- Cao X, Mittelstaedt RA, Pearce MG, Allen BC, Soeteman-Hernandez LG, Johnson GE, Bigger CA, Heflich RH. Quantitative dose-response analysis of ethyl methanesulfonate genotoxicity in adult gpt-delta transgenic mice. Environmental and molecular mutagenesis. 2014;55:385–399. - PubMed
-
- Gocke E, Muller L. In vivo studies in the mouse to define a threshold for the genotoxicity of EMS and ENU. Mutation research. 2009;678:101–107. - PubMed
-
- Marsden DA, Jones DJ, Britton RG, Ognibene T, Ubick E, Johnson GE, Farmer PB, Brown K. Dose-response relationships for N7-(2-hydroxyethyl)guanine induced by low-dose [14C]ethylene oxide: evidence for a novel mechanism of endogenous adduct formation. Cancer research. 2009;69:3052–3059. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

