Network Biomarkers of Bladder Cancer Based on a Genome-Wide Genetic and Epigenetic Network Derived from Next-Generation Sequencing Data
- PMID: 27034531
- PMCID: PMC4789422
- DOI: 10.1155/2016/4149608
Network Biomarkers of Bladder Cancer Based on a Genome-Wide Genetic and Epigenetic Network Derived from Next-Generation Sequencing Data
Abstract
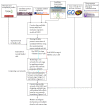
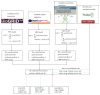
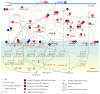
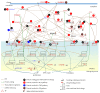
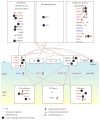
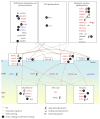
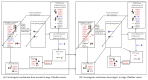
Epigenetic and microRNA (miRNA) regulation are associated with carcinogenesis and the development of cancer. By using the available omics data, including those from next-generation sequencing (NGS), genome-wide methylation profiling, candidate integrated genetic and epigenetic network (IGEN) analysis, and drug response genome-wide microarray analysis, we constructed an IGEN system based on three coupling regression models that characterize protein-protein interaction networks (PPINs), gene regulatory networks (GRNs), miRNA regulatory networks (MRNs), and epigenetic regulatory networks (ERNs). By applying system identification method and principal genome-wide network projection (PGNP) to IGEN analysis, we identified the core network biomarkers to investigate bladder carcinogenic mechanisms and design multiple drug combinations for treating bladder cancer with minimal side-effects. The progression of DNA repair and cell proliferation in stage 1 bladder cancer ultimately results not only in the derepression of miR-200a and miR-200b but also in the regulation of the TNF pathway to metastasis-related genes or proteins, cell proliferation, and DNA repair in stage 4 bladder cancer. We designed a multiple drug combination comprising gefitinib, estradiol, yohimbine, and fulvestrant for treating stage 1 bladder cancer with minimal side-effects, and another multiple drug combination comprising gefitinib, estradiol, chlorpromazine, and LY294002 for treating stage 4 bladder cancer with minimal side-effects.
Figures
Similar articles
-
Investigating core genetic-and-epigenetic cell cycle networks for stemness and carcinogenic mechanisms, and cancer drug design using big database mining and genome-wide next-generation sequencing data.Cell Cycle. 2016 Oct;15(19):2593-2607. doi: 10.1080/15384101.2016.1198862. Epub 2016 Jun 13. Cell Cycle. 2016. PMID: 27295129 Free PMC article.
-
Investigating the mechanism of hepatocellular carcinoma progression by constructing genetic and epigenetic networks using NGS data identification and big database mining method.Oncotarget. 2016 Nov 29;7(48):79453-79473. doi: 10.18632/oncotarget.13100. Oncotarget. 2016. PMID: 27821810 Free PMC article.
-
Constructing an integrated genetic and epigenetic cellular network for whole cellular mechanism using high-throughput next-generation sequencing data.BMC Syst Biol. 2016 Feb 20;10:18. doi: 10.1186/s12918-016-0256-5. BMC Syst Biol. 2016. PMID: 26897165 Free PMC article.
-
MicroRNA expression profiling in bladder cancer: the challenge of next-generation sequencing in tissues and biofluids.Int J Cancer. 2016 May 15;138(10):2334-45. doi: 10.1002/ijc.29895. Epub 2015 Nov 9. Int J Cancer. 2016. PMID: 26489968 Review.
-
Next generation sequencing technologies in cancer diagnostics and therapeutics: A mini review.Cell Mol Biol (Noisy-le-grand). 2015 Oct 30;61(5):91-102. Cell Mol Biol (Noisy-le-grand). 2015. PMID: 26522064 Review.
Cited by
-
Comparing progression molecular mechanisms between lung adenocarcinoma and lung squamous cell carcinoma based on genetic and epigenetic networks: big data mining and genome-wide systems identification.Oncotarget. 2019 Jun 4;10(38):3760-3806. doi: 10.18632/oncotarget.26940. eCollection 2019 Jun 4. Oncotarget. 2019. PMID: 31217907 Free PMC article.
-
Investigating core genetic-and-epigenetic cell cycle networks for stemness and carcinogenic mechanisms, and cancer drug design using big database mining and genome-wide next-generation sequencing data.Cell Cycle. 2016 Oct;15(19):2593-2607. doi: 10.1080/15384101.2016.1198862. Epub 2016 Jun 13. Cell Cycle. 2016. PMID: 27295129 Free PMC article.
-
Genetic-and-Epigenetic Interspecies Networks for Cross-Talk Mechanisms in Human Macrophages and Dendritic Cells during MTB Infection.Front Cell Infect Microbiol. 2016 Oct 18;6:124. doi: 10.3389/fcimb.2016.00124. eCollection 2016. Front Cell Infect Microbiol. 2016. PMID: 27803888 Free PMC article.
-
Investigating the mechanism of hepatocellular carcinoma progression by constructing genetic and epigenetic networks using NGS data identification and big database mining method.Oncotarget. 2016 Nov 29;7(48):79453-79473. doi: 10.18632/oncotarget.13100. Oncotarget. 2016. PMID: 27821810 Free PMC article.
-
Crosstalk of disulfidptosis-related subtypes, establishment of a prognostic signature and immune infiltration characteristics in bladder cancer based on a machine learning survival framework.Front Endocrinol (Lausanne). 2023 Apr 19;14:1180404. doi: 10.3389/fendo.2023.1180404. eCollection 2023. Front Endocrinol (Lausanne). 2023. PMID: 37152941 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

