The degree of intratumor mutational heterogeneity varies by primary tumor sub-site
- PMID: 27034009
- PMCID: PMC5053641
- DOI: 10.18632/oncotarget.8448
The degree of intratumor mutational heterogeneity varies by primary tumor sub-site
Abstract
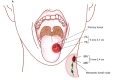
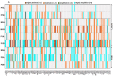
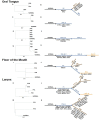
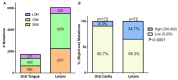
In an era where mutational profiles inform treatment options, it is critical to know the extent to which tumor biopsies represent the molecular profile of the primary and metastatic tumor. Head and neck squamous cell carcinoma (HNSCC) arise primarily in the mucosal lining of oral cavity and oropharynx. Despite aggressive therapy the 5-year survival rate is at 50%. The primary objective of this study is to characterize the degree of intratumor mutational heterogeneity in HNSCC. We used multi-region sequencing of paired primary and metastatic tumor DNA of 24 spatially distinct samples from seven patients with HNSCC of larynx, floor of the mouth (FOM) or oral tongue. Full length, in-depth sequencing of 202 genes implicated in cancer was carried out. Larynx and FOM tumors had more than 69.2% unique SNVs between the paired primary and metastatic lesions. In contrast, the oral tongue HNSCC had only 33.3% unique SNVs across multiple sites. In addition, HNSCC of the oral tongue had fewer mutations than larynx and FOM tumors. These findings were validated on the Affymetrix whole genome 6.0 array platform and were consistent with data from The Cancer Genome Atlas (TCGA). This is the first report demonstrating differences in mutational heterogeneity varying by subsite in HNSCC. The heterogeneity within laryngeal tumor specimens may lead to an underestimation of the genetic abnormalities within tumors and may foster resistance to standard treatment protocols. These findings are relevant to investigators and clinicians developing personalized cancer treatments based on identification of specific mutations in tumor biopsies.
Keywords: HNSCC; HPV; deep sequencing; intratumor heterogeneity; mutation.
Conflict of interest statement
No potential conflicts of interest were identified by any of the authors.
Figures





Similar articles
-
Genomic alterations in head and neck squamous cell carcinoma determined by cancer gene-targeted sequencing.Ann Oncol. 2015 Jun;26(6):1216-1223. doi: 10.1093/annonc/mdv109. Epub 2015 Feb 23. Ann Oncol. 2015. PMID: 25712460 Free PMC article.
-
Identification of mutations in the PYRIN-containing NLR genes (NLRP) in Head and Neck Squamous Cell Carcinoma.PLoS One. 2014 Jan 21;9(1):e85619. doi: 10.1371/journal.pone.0085619. eCollection 2014. PLoS One. 2014. PMID: 24465623 Free PMC article.
-
Variants of EVER1 and EVER2 (TMC6 and TMC8) and human papillomavirus status in patients with mucosal squamous cell carcinoma of the head and neck.Cancer Causes Control. 2016 Jun;27(6):809-15. doi: 10.1007/s10552-016-0749-y. Epub 2016 Apr 20. Cancer Causes Control. 2016. PMID: 27097911
-
Systematic review and individual patient data analysis of pediatric head and neck squamous cell carcinoma: An analysis of 217 cases.Int J Pediatr Otorhinolaryngol. 2017 Jan;92:75-81. doi: 10.1016/j.ijporl.2016.11.005. Epub 2016 Nov 12. Int J Pediatr Otorhinolaryngol. 2017. PMID: 28012539 Review.
-
Head and neck squamous cell carcinoma: Ambiguous human papillomavirus status, elevated p16, and deleted retinoblastoma 1.Head Neck. 2017 Mar;39(3):E34-E39. doi: 10.1002/hed.24604. Epub 2016 Nov 8. Head Neck. 2017. PMID: 27859938 Free PMC article. Review.
Cited by
-
Pathological Bases and Clinical Impact of Intratumor Heterogeneity in Clear Cell Renal Cell Carcinoma.Curr Urol Rep. 2018 Jan 27;19(1):3. doi: 10.1007/s11934-018-0754-7. Curr Urol Rep. 2018. PMID: 29374850 Review.
-
Prognostic Value of Molecular Intratumor Heterogeneity in Primary Oral Cancer and Its Lymph Node Metastases Assessed by Mass Spectrometry Imaging.Molecules. 2022 Aug 25;27(17):5458. doi: 10.3390/molecules27175458. Molecules. 2022. PMID: 36080226 Free PMC article.
-
Metastatic Tumor-in-a-Dish, a Novel Multicellular Organoid to Study Lung Colonization and Predict Therapeutic Response.Cancer Res. 2019 Apr 1;79(7):1681-1695. doi: 10.1158/0008-5472.CAN-18-2602. Epub 2019 Jan 23. Cancer Res. 2019. PMID: 30674533 Free PMC article.
-
De-Escalation Strategies in HPV-Associated Oropharynx Cancer: A Historical Perspective with Future Direction.Cancers (Basel). 2024 Aug 1;16(15):2733. doi: 10.3390/cancers16152733. Cancers (Basel). 2024. PMID: 39123461 Free PMC article. Review.
-
Current Prospects of Molecular Therapeutics in Head and Neck Squamous Cell Carcinoma.Pharmaceut Med. 2019 Aug;33(4):269-289. doi: 10.1007/s40290-019-00288-x. Pharmaceut Med. 2019. PMID: 31933185 Review.
References
-
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA. 2011;61:69–90. - PubMed
-
- Howlader N, N AM, Krapcho M, Garshell J, Neyman N, Altekruse SF, Kosary CL, Yu M, Ruhl J, Tatalovich Z, Cho H, Mariotto A, Lewis DR, Chen HS, Feuer EJ, Cronin KA. SEER Cancer Statistics Review, 1975-2010. National Cancer Institute; Bethesda, MD: 2012. http://seer.cancer.gov/csr/1975_2010/ based on November 2012 SEER data submission.
-
- Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, Fakhry C, Xie TX, Zhang J, Wang J, Zhang N, El-Naggar AK, Jasser SA, Weinstein JN, Trevino L, Drummond JA, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

