Early ERK1/2 activation promotes DRP1-dependent mitochondrial fission necessary for cell reprogramming
- PMID: 27030341
- PMCID: PMC4821885
- DOI: 10.1038/ncomms11124
Early ERK1/2 activation promotes DRP1-dependent mitochondrial fission necessary for cell reprogramming
Abstract
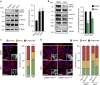
During the process of reprogramming to induced pluripotent stem (iPS) cells, somatic cells switch from oxidative to glycolytic metabolism, a transition associated with profound mitochondrial reorganization. Neither the importance of mitochondrial remodelling for cell reprogramming, nor the molecular mechanisms controlling this process are well understood. Here, we show that an early wave of mitochondrial fragmentation occurs upon expression of reprogramming factors. Reprogramming-induced mitochondrial fission is associated with a minor decrease in mitochondrial mass but not with mitophagy. The pro-fission factor Drp1 is phosphorylated early in reprogramming, and its knockdown and inhibition impairs both mitochondrial fragmentation and generation of iPS cell colonies. Drp1 phosphorylation depends on Erk activation in early reprogramming, which occurs, at least in part, due to downregulation of the MAP kinase phosphatase Dusp6. Taken together, our data indicate that mitochondrial fission controlled by an Erk-Drp1 axis constitutes an early and necessary step in the reprogramming process to pluripotency.
Figures

Similar articles
-
Drp1 is dispensable for mitochondria biogenesis in induction to pluripotency but required for differentiation of embryonic stem cells.Stem Cells Dev. 2014 Oct 15;23(20):2422-34. doi: 10.1089/scd.2014.0059. Epub 2014 Aug 4. Stem Cells Dev. 2014. PMID: 24937776
-
c-MYC independent nuclear reprogramming favors cardiogenic potential of induced pluripotent stem cells.J Cardiovasc Transl Res. 2010 Feb;3(1):13-23. doi: 10.1007/s12265-009-9150-5. J Cardiovasc Transl Res. 2010. PMID: 20221419 Free PMC article.
-
SIRT2 regulates mitochondrial dynamics and reprogramming via MEK1-ERK-DRP1 and AKT1-DRP1 axes.Cell Rep. 2021 Dec 28;37(13):110155. doi: 10.1016/j.celrep.2021.110155. Cell Rep. 2021. PMID: 34965411 Free PMC article.
-
The genetics of induced pluripotency.Reproduction. 2010 Jan;139(1):35-44. doi: 10.1530/REP-09-0024. Reproduction. 2010. PMID: 19605512 Review.
-
Mechanism of Induction: Induced Pluripotent Stem Cells (iPSCs).J Stem Cells. 2015;10(1):43-62. J Stem Cells. 2015. PMID: 26665937 Review.
Cited by
-
Dynamin-Related Protein 1 Binding Partners MiD49 and MiD51 Increased Mitochondrial Fission In Vitro and Atherosclerosis in High-Fat-Diet-Fed ApoE-/- Mice.Int J Mol Sci. 2023 Dec 23;25(1):244. doi: 10.3390/ijms25010244. Int J Mol Sci. 2023. PMID: 38203413 Free PMC article.
-
MYO1F Regulates IgE and MRGPRX2-Dependent Mast Cell Exocytosis.J Immunol. 2021 May 15;206(10):2277-2289. doi: 10.4049/jimmunol.2001211. Epub 2021 May 3. J Immunol. 2021. PMID: 33941653 Free PMC article.
-
CDDO-Me Selectively Attenuates CA1 Neuronal Death Induced by Status Epilepticus via Facilitating Mitochondrial Fission Independent of LONP1.Cells. 2019 Aug 5;8(8):833. doi: 10.3390/cells8080833. Cells. 2019. PMID: 31387295 Free PMC article.
-
Metabolic Plasticity in Chemotherapy Resistance.Front Oncol. 2020 Mar 6;10:281. doi: 10.3389/fonc.2020.00281. eCollection 2020. Front Oncol. 2020. PMID: 32211323 Free PMC article. Review.
-
HIF-1α Affects the Neural Stem Cell Differentiation of Human Induced Pluripotent Stem Cells via MFN2-Mediated Wnt/β-Catenin Signaling.Front Cell Dev Biol. 2021 Jun 21;9:671704. doi: 10.3389/fcell.2021.671704. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34235146 Free PMC article.
References
-
- Takahashi K. & Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006). - PubMed
-
- Okita K., Ichisaka T. & Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature 448, 313–317 (2007). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

