Human Cytomegalovirus Infection Upregulates the Mitochondrial Transcription and Translation Machineries
- PMID: 27025248
- PMCID: PMC4807356
- DOI: 10.1128/mBio.00029-16
Human Cytomegalovirus Infection Upregulates the Mitochondrial Transcription and Translation Machineries
Abstract
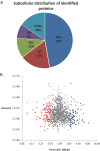
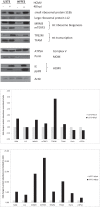
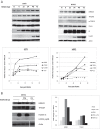
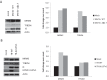
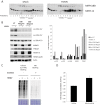
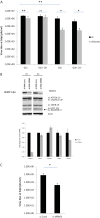
Infection with human cytomegalovirus (HCMV) profoundly affects cellular metabolism. Like in tumor cells, HCMV infection increases glycolysis, and glucose carbon is shifted from the mitochondrial tricarboxylic acid cycle to the biosynthesis of fatty acids. However, unlike in many tumor cells, where aerobic glycolysis is accompanied by suppression of mitochondrial oxidative phosphorylation, HCMV induces mitochondrial biogenesis and respiration. Here, we affinity purified mitochondria and used quantitative mass spectrometry to determine how the mitochondrial proteome changes upon HCMV infection. We found that the mitochondrial transcription and translation systems are induced early during the viral replication cycle. Specifically, proteins involved in biogenesis of the mitochondrial ribosome were highly upregulated by HCMV infection. Inhibition of mitochondrial translation with chloramphenicol or knockdown of HCMV-induced ribosome biogenesis factor MRM3 abolished the HCMV-mediated increase in mitochondrially encoded proteins and significantly impaired viral growth under bioenergetically restricting conditions. Our findings demonstrate how HCMV manipulates mitochondrial biogenesis to support its replication.
Importance: Human cytomegalovirus (HCMV), a betaherpesvirus, is a leading cause of morbidity and mortality during congenital infection and among immunosuppressed individuals. HCMV infection significantly changes cellular metabolism. Akin to tumor cells, in HCMV-infected cells, glycolysis is increased and glucose carbon is shifted from the tricarboxylic acid cycle to fatty acid biosynthesis. However, unlike in tumor cells, HCMV induces mitochondrial biogenesis even under aerobic glycolysis. Here, we have affinity purified mitochondria and used quantitative mass spectrometry to determine how the mitochondrial proteome changes upon HCMV infection. We find that the mitochondrial transcription and translation systems are induced early during the viral replication cycle. Specifically, proteins involved in biogenesis of the mitochondrial ribosome were highly upregulated by HCMV infection. Inhibition of mitochondrial translation with chloramphenicol or knockdown of HCMV-induced ribosome biogenesis factor MRM3 abolished the HCMV-mediated increase in mitochondrially encoded proteins and significantly impaired viral growth. Our findings demonstrate how HCMV manipulates mitochondrial biogenesis to support its replication.
Copyright © 2016 Karniely et al.
Figures
Similar articles
-
Human Cytomegalovirus Alters Host Cell Mitochondrial Function during Acute Infection.J Virol. 2020 Jan 6;94(2):e01183-19. doi: 10.1128/JVI.01183-19. Print 2020 Jan 6. J Virol. 2020. PMID: 31694945 Free PMC article.
-
Nitric Oxide Circumvents Virus-Mediated Metabolic Regulation during Human Cytomegalovirus Infection.mBio. 2020 Dec 15;11(6):e02630-20. doi: 10.1128/mBio.02630-20. mBio. 2020. PMID: 33323506 Free PMC article.
-
Human Cytomegalovirus pUL37x1 Is Important for Remodeling of Host Lipid Metabolism.J Virol. 2019 Oct 15;93(21):e00843-19. doi: 10.1128/JVI.00843-19. Print 2019 Nov 1. J Virol. 2019. PMID: 31391267 Free PMC article.
-
Viral effects on metabolism: changes in glucose and glutamine utilization during human cytomegalovirus infection.Trends Microbiol. 2011 Jul;19(7):360-7. doi: 10.1016/j.tim.2011.04.002. Epub 2011 May 12. Trends Microbiol. 2011. PMID: 21570293 Free PMC article. Review.
-
Human Cytomegalovirus Strategies to Maintain and Promote mRNA Translation.Viruses. 2016 Apr 13;8(4):97. doi: 10.3390/v8040097. Viruses. 2016. PMID: 27089357 Free PMC article. Review.
Cited by
-
Host Mitochondrial Requirements of Cytomegalovirus Replication.Curr Clin Microbiol Rep. 2020 Dec;7(4):115-123. doi: 10.1007/s40588-020-00153-5. Epub 2020 Sep 30. Curr Clin Microbiol Rep. 2020. PMID: 33816061 Free PMC article.
-
Transcriptome Analysis on Hepatopancreas Reveals the Metabolic Dysregulation Caused by Vibrio parahaemolyticus Infection in Litopenaeus vannamei.Biology (Basel). 2023 Mar 9;12(3):417. doi: 10.3390/biology12030417. Biology (Basel). 2023. PMID: 36979109 Free PMC article.
-
Mitochondrial Oxidative Phosphorylation in Viral Infections.Viruses. 2023 Dec 4;15(12):2380. doi: 10.3390/v15122380. Viruses. 2023. PMID: 38140621 Free PMC article. Review.
-
Meal for Two: Human Cytomegalovirus-Induced Activation of Cellular Metabolism.Viruses. 2019 Mar 19;11(3):273. doi: 10.3390/v11030273. Viruses. 2019. PMID: 30893762 Free PMC article. Review.
-
Human Cytomegalovirus Alters Host Cell Mitochondrial Function during Acute Infection.J Virol. 2020 Jan 6;94(2):e01183-19. doi: 10.1128/JVI.01183-19. Print 2020 Jan 6. J Virol. 2020. PMID: 31694945 Free PMC article.
References
-
- Boppana SB, Britt WJ. 2013. Synopsis of clinical aspects of human cytomegalovirus disease, p 1–25. In Reddehase MJ. (ed), Cytomegaloviruses: from molecular pathogenesis to intervention. Caister Academic Press, Norfolk, United Kingdom.
-
- Rabinowitz JD, Shenk T, Reddehase M. 2013. Human cytomegalovirus metabolomics, p 59–67. In Reddehase MJ. (ed), Cytomegaloviruses: from molecular pathogenesis to intervention. Caister Academic Press, Norfolk, United Kingdom.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
