miR-isomiRExp: a web-server for the analysis of expression of miRNA at the miRNA/isomiR levels
- PMID: 27009551
- PMCID: PMC4806314
- DOI: 10.1038/srep23700
miR-isomiRExp: a web-server for the analysis of expression of miRNA at the miRNA/isomiR levels
Abstract
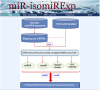
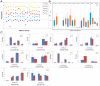
MicroRNA (miRNA) locus has been found that can generate a series of varied isomiR sequences. Most studies always focus on determining miRNA level, however, the canonical miRNA sequence is only a specific member in the multiple isomiRs. Some studies have shown that isomiR sequences play versatile roles in biological progress, and the analysis and research should be simultaneously performed at the miRNA/isomiR levels. Based on the biological characteristics of miRNA and isomiR, we developed miR-isomiRExp to analyze expression pattern of miRNA at the miRNA/isomiR levels, provide insights into tracking miRNA/isomiR maturation and processing mechanisms, and reveal functional characteristics of miRNA/isomiR. Simultaneously, we also performed expression analysis of specific human diseases using public small RNA sequencing datasets based on the analysis platform, which may help in surveying the potential deregulated miRNA/isomiR expression profiles, especially sequence and function-related isomiRs for further interaction analysis and study. The miR-isomiRExp platform provides miRNA/isomiR expression patterns and more information to study deregulated miRNA loci and detailed isomiR sequences. This comprehensive analysis will enrich experimental miRNA studies. miR-isomiRExp is available at http://mirisomirexp.aliapp.com.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

Similar articles
-
mi-IsoNet: systems-scale microRNA landscape reveals rampant isoform-mediated gain of target interaction diversity and signaling specificity.Brief Bioinform. 2021 Sep 2;22(5):bbab091. doi: 10.1093/bib/bbab091. Brief Bioinform. 2021. PMID: 33855356 Free PMC article.
-
A Comprehensive Approach to Sequence-oriented IsomiR annotation (CASMIR): demonstration with IsomiR profiling in colorectal neoplasia.BMC Genomics. 2018 May 25;19(1):401. doi: 10.1186/s12864-018-4794-7. BMC Genomics. 2018. PMID: 29801434 Free PMC article.
-
Comprehensive expression analysis of miRNA in breast cancer at the miRNA and isomiR levels.Gene. 2015 Feb 25;557(2):195-200. doi: 10.1016/j.gene.2014.12.030. Epub 2014 Dec 16. Gene. 2015. PMID: 25523096
-
A challenge for miRNA: multiple isomiRs in miRNAomics.Gene. 2014 Jul 1;544(1):1-7. doi: 10.1016/j.gene.2014.04.039. Epub 2014 Apr 21. Gene. 2014. PMID: 24768184 Review.
-
Identification and validation of plant miRNA from NGS data-an experimental approach.Brief Funct Genomics. 2019 Feb 14;18(1):13-22. doi: 10.1093/bfgp/ely034. Brief Funct Genomics. 2019. PMID: 30335137 Review.
Cited by
-
Evidence for plant-derived xenomiRs based on a large-scale analysis of public small RNA sequencing data from human samples.PLoS One. 2018 Jun 27;13(6):e0187519. doi: 10.1371/journal.pone.0187519. eCollection 2018. PLoS One. 2018. PMID: 29949574 Free PMC article.
-
Small RNAs in Circulating Exosomes of Cancer Patients: A Minireview.High Throughput. 2017 Oct 6;6(4):13. doi: 10.3390/ht6040013. High Throughput. 2017. PMID: 29485611 Free PMC article. Review.
-
Comparative Mucous miRomics in Cynoglossus semilaevis Related to Vibrio harveyi Caused Infection.Mar Biotechnol (NY). 2021 Oct;23(5):766-776. doi: 10.1007/s10126-021-10062-3. Epub 2021 Sep 3. Mar Biotechnol (NY). 2021. PMID: 34480240
-
Genome-wide miRNA profiling reinforces the importance of miR-9 in human papillomavirus associated oral and oropharyngeal head and neck cancer.Sci Rep. 2019 Feb 19;9(1):2306. doi: 10.1038/s41598-019-38797-z. Sci Rep. 2019. PMID: 30783190 Free PMC article.
-
Web-based NGS data analysis using miRMaster: a large-scale meta-analysis of human miRNAs.Nucleic Acids Res. 2017 Sep 6;45(15):8731-8744. doi: 10.1093/nar/gkx595. Nucleic Acids Res. 2017. PMID: 28911107 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

