The novel RacE-binding protein GflB sharpens Ras activity at the leading edge of migrating cells
- PMID: 27009206
- PMCID: PMC4865317
- DOI: 10.1091/mbc.E15-11-0796
The novel RacE-binding protein GflB sharpens Ras activity at the leading edge of migrating cells
Abstract
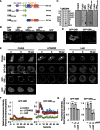
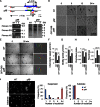
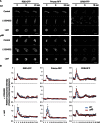
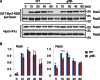
Directional sensing, a process in which cells convert an external chemical gradient into internal signaling events, is essential in chemotaxis. We previously showed that a Rho GTPase, RacE, regulates gradient sensing in Dictyostelium cells. Here, using affinity purification and mass spectrometry, we identify a novel RacE-binding protein, GflB, which contains a Ras GEF domain and a Rho GAP domain. Using biochemical and gene knockout approaches, we show that GflB balances the activation of Ras and Rho GTPases, which enables cells to precisely orient signaling events toward higher concentrations of chemoattractants. Furthermore, we find that GflB is located at the leading edge of migrating cells, and this localization is regulated by the actin cytoskeleton and phosphatidylserine. Our findings provide a new molecular mechanism that connects directional sensing and morphological polarization.
© 2016 Senoo et al. This article is distributed by The American Society for Cell Biology under license from the author(s). Two months after publication it is available to the public under an Attribution–Noncommercial–Share Alike 3.0 Unported Creative Commons License (http://creativecommons.org/licenses/by-nc-sa/3.0).
Figures

Similar articles
-
The F-actin-binding RapGEF GflB is required for efficient macropinocytosis in Dictyostelium.J Cell Sci. 2017 Sep 15;130(18):3158-3172. doi: 10.1242/jcs.194126. Epub 2017 Aug 4. J Cell Sci. 2017. PMID: 28778987
-
A Gα-Stimulated RapGEF Is a Receptor-Proximal Regulator of Dictyostelium Chemotaxis.Dev Cell. 2016 Jun 6;37(5):458-72. doi: 10.1016/j.devcel.2016.05.001. Epub 2016 May 26. Dev Cell. 2016. PMID: 27237792
-
GPCR-controlled membrane recruitment of negative regulator C2GAP1 locally inhibits Ras signaling for adaptation and long-range chemotaxis.Proc Natl Acad Sci U S A. 2017 Nov 21;114(47):E10092-E10101. doi: 10.1073/pnas.1703208114. Epub 2017 Nov 6. Proc Natl Acad Sci U S A. 2017. PMID: 29109256 Free PMC article.
-
Rho Signaling in Dictyostelium discoideum.Int Rev Cell Mol Biol. 2016;322:61-181. doi: 10.1016/bs.ircmb.2015.10.004. Epub 2015 Dec 23. Int Rev Cell Mol Biol. 2016. PMID: 26940518 Review.
-
Leading the way: Directional sensing through phosphatidylinositol 3-kinase and other signaling pathways.J Cell Sci. 2003 Sep 1;116(Pt 17):3471-8. doi: 10.1242/jcs.00703. J Cell Sci. 2003. PMID: 12893811 Review.
Cited by
-
The nucleus does not significantly affect the migratory trajectories of amoeba in two-dimensional environments.Sci Rep. 2019 Nov 8;9(1):16369. doi: 10.1038/s41598-019-52716-2. Sci Rep. 2019. PMID: 31704992 Free PMC article.
-
Excitable networks controlling cell migration during development and disease.Semin Cell Dev Biol. 2020 Apr;100:133-142. doi: 10.1016/j.semcdb.2019.11.001. Epub 2019 Dec 10. Semin Cell Dev Biol. 2020. PMID: 31836289 Free PMC article. Review.
-
Coupled excitable Ras and F-actin activation mediates spontaneous pseudopod formation and directed cell movement.Mol Biol Cell. 2017 Apr 1;28(7):922-934. doi: 10.1091/mbc.E16-10-0733. Epub 2017 Feb 1. Mol Biol Cell. 2017. PMID: 28148648 Free PMC article.
-
Hetero-oligomerization of Rho and Ras GTPases Connects GPCR Activation to mTORC2-AKT Signaling.Cell Rep. 2020 Nov 24;33(8):108427. doi: 10.1016/j.celrep.2020.108427. Cell Rep. 2020. PMID: 33238110 Free PMC article.
-
Functional integrity of the contractile actin cortex is safeguarded by multiple Diaphanous-related formins.Proc Natl Acad Sci U S A. 2019 Feb 26;116(9):3594-3603. doi: 10.1073/pnas.1821638116. Epub 2019 Feb 11. Proc Natl Acad Sci U S A. 2019. PMID: 30808751 Free PMC article.
References
-
- Aman A, Piotrowski T. Cell migration during morphogenesis. Dev Biol. 2010;341:20–33. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

