Identification of Genes That Modulate Susceptibility to Formaldehyde and Imatinib by Functional Genomic Screening in Human Haploid KBM7 Cells
- PMID: 27008852
- PMCID: PMC4914802
- DOI: 10.1093/toxsci/kfw032
Identification of Genes That Modulate Susceptibility to Formaldehyde and Imatinib by Functional Genomic Screening in Human Haploid KBM7 Cells
Erratum in
-
Identification of Genes That Modulate Susceptibility to Formaldehyde and Imatinib by Functional Genomic Screening in Human Haploid KBM7 Cells.Toxicol Sci. 2016 Nov;154(1):194. doi: 10.1093/toxsci/kfw183. Toxicol Sci. 2016. PMID: 27794141 Free PMC article. No abstract available.
Abstract
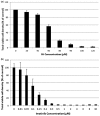
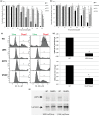
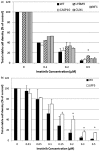
Though current functional genomic screening systems are useful for investigating human susceptibility to chemical toxicity, they have limitations. Well-established, high-throughput yeast mutant screens identify only evolutionarily conserved processes. RNA interference can be applied in human cells but is limited by incomplete gene knockout and off-target effects. Human haploid cell screening is advantageous as it requires knockdown of only a single copy of each gene. A human haploid cell mutant library (KBM7-Mu), derived from a chronic myeloid leukemia (CML) patient, was recently developed and has been used to identify genes that modulate sensitivity to infectious agents and pharmaceutical drugs. Here, we sought to improve the KBM7-Mu screening process to enable efficient screening of environmental chemicals. We developed a semi-solid medium based screening approach that cultures individual mutant colonies from chemically resistant cells, faster (by 2-3 weeks) and with less labor than the original liquid medium-based approach. As proof of principle, we identified genetic mutants that confer resistance to the carcinogen formaldehyde (FA, 12 genes, 18 hits) and the CML chemotherapeutic agent imatinib (6 genes, 13 hits). Validation experiments conducted on KBM7 mutants lacking each of the 18 genes confirmed resistance of 6 FA mutants (CTC1, FCRLA, GOT1, LPR5, M1AP, and MAP2K5) and 1 imatinib-resistant mutant (LYRM9). Despite the improvements to the method, it remains technically challenging to limit false positive findings. Nonetheless, our findings demonstrate the broad applicability of this optimized haploid approach to screen toxic chemicals to identify novel susceptibility genes and gain insight into potential mechanisms of toxicity.
Keywords: KBM7; formaldehyde; functional genomics; haploid; imatinib..
© The Author 2016. Published by Oxford University Press on behalf of the Society of Toxicology. All rights reserved. For Permissions, please e-mail: journals.permissions@oup.com.
Figures
Similar articles
-
Application of human haploid cell genetic screening model in identifying the genes required for resistance to environmental toxicants: Chlorpyrifos as a case study.J Pharmacol Toxicol Methods. 2015 Nov-Dec;76:76-82. doi: 10.1016/j.vascn.2015.08.154. Epub 2015 Aug 20. J Pharmacol Toxicol Methods. 2015. PMID: 26299976 Free PMC article.
-
HS-543 induces apoptosis of Imatinib-resistant chronic myelogenous leukemia with T315I mutation.Oncotarget. 2015 Jan 30;6(3):1507-18. doi: 10.18632/oncotarget.2837. Oncotarget. 2015. PMID: 25483100 Free PMC article.
-
Revealing genome-wide mRNA and microRNA expression patterns in leukemic cells highlighted "hsa-miR-2278" as a tumor suppressor for regain of chemotherapeutic imatinib response due to targeting STAT5A.Tumour Biol. 2015 Sep;36(10):7915-27. doi: 10.1007/s13277-015-3509-9. Epub 2015 May 8. Tumour Biol. 2015. PMID: 25953263
-
Sensitivity of imatinib-resistant T315I BCR-ABL CML to a synergistic combination of ponatinib and forskolin treatment.Tumour Biol. 2016 Sep;37(9):12643-12654. doi: 10.1007/s13277-016-5179-7. Epub 2016 Jul 21. Tumour Biol. 2016. PMID: 27444277 Free PMC article.
-
Application of toxicogenomic profiling to evaluate effects of benzene and formaldehyde: from yeast to human.Ann N Y Acad Sci. 2014 Mar;1310(1):74-83. doi: 10.1111/nyas.12382. Epub 2014 Feb 26. Ann N Y Acad Sci. 2014. PMID: 24571325 Free PMC article. Review.
Cited by
-
Applying genome-wide CRISPR to identify known and novel genes and pathways that modulate formaldehyde toxicity.Chemosphere. 2021 Apr;269:128701. doi: 10.1016/j.chemosphere.2020.128701. Epub 2020 Oct 22. Chemosphere. 2021. PMID: 33189395 Free PMC article.
-
Bridging the Gap in Cancer Research: Sulfur Metabolism of Leukemic Cells with a Focus on L-Cysteine Metabolism and Hydrogen Sulfide-Producing Enzymes.Biomolecules. 2024 Jun 24;14(7):746. doi: 10.3390/biom14070746. Biomolecules. 2024. PMID: 39062461 Free PMC article. Review.
-
SIRT3 Expression Predicts Overall Survival and Neoadjuvant Chemosensitivity in Triple-Negative Breast Cancer.Cancer Manag Res. 2024 Mar 8;16:137-150. doi: 10.2147/CMAR.S445248. eCollection 2024. Cancer Manag Res. 2024. PMID: 38476973 Free PMC article.
-
New tools for old drugs: Functional genetic screens to optimize current chemotherapy.Drug Resist Updat. 2018 Jan;36:30-46. doi: 10.1016/j.drup.2018.01.001. Epub 2018 Jan 12. Drug Resist Updat. 2018. PMID: 29499836 Free PMC article. Review.
-
Functional Toxicogenomic Profiling Expands Insight into Modulators of Formaldehyde Toxicity in Yeast.Front Genet. 2016 Nov 17;7:200. doi: 10.3389/fgene.2016.00200. eCollection 2016. Front Genet. 2016. PMID: 27909446 Free PMC article.
References
-
- Andersen M. E., Clewell H. J., III, Bermudez E., Dodd D. E., Willson G. A., Campbell J. L., Thomas R. S. (2010). Formaldehyde: Integrating dosimetry, cytotoxicity, and genomics to understand dose-dependent transitions for an endogenous compound. Toxicol. Sci. 118, 716–731. - PubMed
-
- Apperley J. F. (2007). Part I: Mechanisms of resistance to imatinib in chronic myeloid leukaemia. Lancet Oncol. 8, 1018–1029. - PubMed
-
- Balabanov S., Braig M., Brummendorf T. H. (2014). Current aspects in resistance against tyrosine kinase inhibitors in chronic myelogenous leukemia. Drug Discov. Today Technol. 11, 89–99. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

