PPARγ neddylation essential for adipogenesis is a potential target for treating obesity
- PMID: 26990658
- PMCID: PMC4947677
- DOI: 10.1038/cdd.2016.6
PPARγ neddylation essential for adipogenesis is a potential target for treating obesity
Abstract
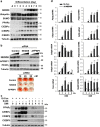
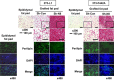
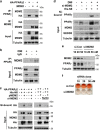
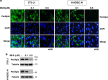
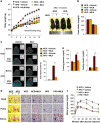
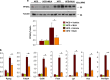
The preadipocyte-to-adipocyte differentiation (adipogenesis) is a key process in fat mass increase and thus it is regarded as a compelling target for preventing or treating obesity. Of adipogenic hormone receptors, peroxisome proliferator-activated receptor gamma (PPARγ) has crucial roles in adipogenesis and lipid accumulation within adipocytes. Here we demonstrate that the NEDD8 (neuronal precursor cell expressed, developmentally downregulated 8)-based post-translation modification (neddylation) of PPARγ is essential for adipogenesis. During adipogenesis, NEDD8 is robustly induced in preadipocytes and conjugates with PPARγ, leading to PPARγ stabilization. When the neddylation process was blocked by NEDD8-targeting siRNAs (or viral vectors) or an inhibitor MLN4924, adipocyte differentiation and fat tissue development were substantially impaired. We also demonstrate that MLN4924 effectively prevents the high-fat diet-induced obesity and glucose intolerance in mice. This study provides a better understanding of how the PPARγ signaling pathway starts and lasts during adipogenesis and a potential anti-obesity strategy that targets the neddylation of PPARγ.
Figures







Similar articles
-
Neddylation and Its Target Cullin 3 Are Essential for Adipocyte Differentiation.Cells. 2024 Oct 5;13(19):1654. doi: 10.3390/cells13191654. Cells. 2024. PMID: 39404417 Free PMC article.
-
Promotion of adipogenesis by 15-(S)-hydroxyeicosatetraenoic acid.Prostaglandins Other Lipid Mediat. 2016 Mar;123:1-8. doi: 10.1016/j.prostaglandins.2016.02.001. Epub 2016 Feb 22. Prostaglandins Other Lipid Mediat. 2016. PMID: 26905195
-
Suppressive effects of saponin-enriched extracts from quinoa on 3T3-L1 adipocyte differentiation.Food Funct. 2015 Oct;6(10):3282-90. doi: 10.1039/c5fo00716j. Food Funct. 2015. PMID: 26242624
-
Molecular Regulation of Adipogenesis and Potential Anti-Adipogenic Bioactive Molecules.Int J Mol Sci. 2016 Jan 19;17(1):124. doi: 10.3390/ijms17010124. Int J Mol Sci. 2016. PMID: 26797605 Free PMC article. Review.
-
Potential of Natural Products in the Inhibition of Adipogenesis through Regulation of PPARγ Expression and/or Its Transcriptional Activity.Molecules. 2016 Sep 23;21(10):1278. doi: 10.3390/molecules21101278. Molecules. 2016. PMID: 27669202 Free PMC article. Review.
Cited by
-
Neddylation Regulates Class IIa and III Histone Deacetylases to Mediate Myoblast Differentiation.Int J Mol Sci. 2021 Sep 1;22(17):9509. doi: 10.3390/ijms22179509. Int J Mol Sci. 2021. PMID: 34502418 Free PMC article.
-
Integrated analysis reveals the regulatory mechanism of the neddylation inhibitor MLN4924 on the metabolic dysregulation in rabbit granulosa cells.BMC Genomics. 2024 Mar 6;25(1):254. doi: 10.1186/s12864-024-10118-3. BMC Genomics. 2024. PMID: 38448814 Free PMC article.
-
Prevalence of hemorrhagic fever with renal syndrome in Qingdao City, China, 2010-2014.Sci Rep. 2016 Oct 27;6:36081. doi: 10.1038/srep36081. Sci Rep. 2016. PMID: 27786303 Free PMC article.
-
Neddylation mediates ventricular chamber maturation through repression of Hippo signaling.Proc Natl Acad Sci U S A. 2018 Apr 24;115(17):E4101-E4110. doi: 10.1073/pnas.1719309115. Epub 2018 Apr 9. Proc Natl Acad Sci U S A. 2018. PMID: 29632206 Free PMC article.
-
Autologous Fat Grafting-A Panacea for Scar Tissue Therapy?Cells. 2024 Aug 20;13(16):1384. doi: 10.3390/cells13161384. Cells. 2024. PMID: 39195271 Free PMC article. Review.
References
-
- Kopelman PG. Obesity as a medical problem. Nature 2000; 404: 635–643. - PubMed
-
- Spalding KL, Arner E, Westermark PO, Bernard S, Buchholz BA, Bergmann O et al. Dynamics of fat cell turnover in humans. Nature 2008; 453: 783–787. - PubMed
-
- Camp HS, Ren D, Leff T. Adipogenesis and fat-cell function in obesity and diabetes. Trends Mol Med 2002; 9: 442–447. - PubMed
-
- Cao Z, Umek RM, McKnight SL. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev 1991; 5: 1538–1552. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

