Signaling via the NFκB system
- PMID: 26990581
- PMCID: PMC8363188
- DOI: 10.1002/wsbm.1331
Signaling via the NFκB system
Abstract
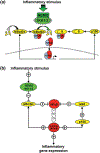
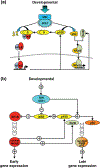
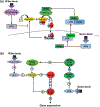
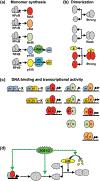
The nuclear factor kappa B (NFκB) family of transcription factors is a key regulator of immune development, immune responses, inflammation, and cancer. The NFκB signaling system (defined by the interactions between NFκB dimers, IκB regulators, and IKK complexes) is responsive to a number of stimuli, and upon ligand-receptor engagement, distinct cellular outcomes, appropriate to the specific signal received, are set into motion. After almost three decades of study, many signaling mechanisms are well understood, rendering them amenable to mathematical modeling, which can reveal deeper insights about the regulatory design principles. While other reviews have focused on upstream, receptor proximal signaling (Hayden MS, Ghosh S. Signaling to NF-κB. Genes Dev 2004, 18:2195-2224; Verstrepen L, Bekaert T, Chau TL, Tavernier J, Chariot A, Beyaert R. TLR-4, IL-1R and TNF-R signaling to NF-κB: variations on a common theme. Cell Mol Life Sci 2008, 65:2964-2978), and advances through computational modeling (Basak S, Behar M, Hoffmann A. Lessons from mathematically modeling the NF-κB pathway. Immunol Rev 2012, 246:221-238; Williams R, Timmis J, Qwarnstrom E. Computational models of the NF-KB signalling pathway. Computation 2014, 2:131), in this review we aim to summarize the current understanding of the NFκB signaling system itself, the molecular mechanisms, and systems properties that are key to its diverse biological functions, and we discuss remaining questions in the field. WIREs Syst Biol Med 2016, 8:227-241. doi: 10.1002/wsbm.1331 For further resources related to this article, please visit the WIREs website.
© 2016 Wiley Periodicals, Inc.
Conflict of interest statement
Conflict of interest: The authors have declared no conflicts of interest for this article.
Figures

Similar articles
-
Protein Kinase-Mediated Decision Between the Life and Death.Adv Exp Med Biol. 2021;1275:1-33. doi: 10.1007/978-3-030-49844-3_1. Adv Exp Med Biol. 2021. PMID: 33539010
-
Regulation and function of IKK and IKK-related kinases.Sci STKE. 2006 Oct 17;2006(357):re13. doi: 10.1126/stke.3572006re13. Sci STKE. 2006. PMID: 17047224 Review.
-
Classical NF-κB activation impairs skeletal muscle oxidative phenotype by reducing IKK-α expression.Biochim Biophys Acta. 2014 Feb;1842(2):175-85. doi: 10.1016/j.bbadis.2013.11.001. Epub 2013 Nov 8. Biochim Biophys Acta. 2014. PMID: 24215713
-
A single NFκB system for both canonical and non-canonical signaling.Cell Res. 2011 Jan;21(1):86-102. doi: 10.1038/cr.2010.161. Epub 2010 Nov 23. Cell Res. 2011. PMID: 21102550 Free PMC article. Review.
-
Recent advances towards understanding redox mechanisms in the activation of nuclear factor kappaB.Free Radic Biol Med. 2000 May 1;28(9):1317-27. doi: 10.1016/s0891-5849(00)00218-5. Free Radic Biol Med. 2000. PMID: 10924851 Review.
Cited by
-
LXR agonist inhibits inflammation through regulating MyD88 mRNA alternative splicing.Front Pharmacol. 2022 Oct 14;13:973612. doi: 10.3389/fphar.2022.973612. eCollection 2022. Front Pharmacol. 2022. PMID: 36313296 Free PMC article.
-
Whole transcriptome sequencing reveals neutrophils' transcriptional landscape associated with active tuberculosis.Front Immunol. 2022 Aug 18;13:954221. doi: 10.3389/fimmu.2022.954221. eCollection 2022. Front Immunol. 2022. PMID: 36059536 Free PMC article.
-
Lithium and Atypical Antipsychotics: The Possible WNT/β Pathway Target in Glaucoma.Biomedicines. 2021 Apr 26;9(5):473. doi: 10.3390/biomedicines9050473. Biomedicines. 2021. PMID: 33925885 Free PMC article. Review.
-
The Emerging Therapeutic Potential of Nitro Fatty Acids and Other Michael Acceptor-Containing Drugs for the Treatment of Inflammation and Cancer.Front Pharmacol. 2020 Sep 3;11:1297. doi: 10.3389/fphar.2020.01297. eCollection 2020. Front Pharmacol. 2020. PMID: 33013366 Free PMC article. Review.
-
TCM and related active compounds in the treatment of gout: the regulation of signaling pathway and urate transporter.Front Pharmacol. 2023 Nov 29;14:1275974. doi: 10.3389/fphar.2023.1275974. eCollection 2023. Front Pharmacol. 2023. PMID: 38094893 Free PMC article. Review.
References
-
- Gerondakis S, Grumont R, Gugasyan R, Wong L, Isomura I, Ho W, Banerjee A. Unravelling the complexities of the NF-κB signalling pathway using mouse knockout and transgenic models. Oncogene 2006, 25:6781–6799. - PubMed
-
- Vallabhapurapu S, Karin M. Regulation and function of NF-κB transcription factors in the immune system. Annu Rev Immunol 2009, 27:693–733. - PubMed
-
- Hoffmann A, Baltimore D. Circuitry of nuclear factor κB signaling. Immunol Rev 2006, 210:171–186. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

