RNA-dependent RNA polymerase 1 in potato (Solanum tuberosum) and its relationship to other plant RNA-dependent RNA polymerases
- PMID: 26979928
- PMCID: PMC4793286
- DOI: 10.1038/srep23082
RNA-dependent RNA polymerase 1 in potato (Solanum tuberosum) and its relationship to other plant RNA-dependent RNA polymerases
Abstract
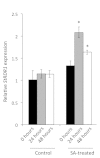
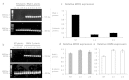
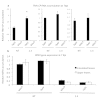
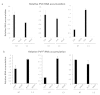
Cellular RNA-dependent RNA polymerases (RDRs) catalyze synthesis of double-stranded RNAs that can serve to initiate or amplify RNA silencing. Arabidopsis thaliana has six RDR genes; RDRs 1, 2 and 6 have roles in anti-viral RNA silencing. RDR6 is constitutively expressed but RDR1 expression is elevated following plant treatment with defensive phytohormones. RDR1 also contributes to basal virus resistance. RDR1 has been studied in several species including A. thaliana, tobacco (Nicotiana tabacum), N. benthamiana, N. attenuata and tomato (Solanum lycopersicum) but not to our knowledge in potato (S. tuberosum). StRDR1 was identified and shown to be salicylic acid-responsive. StRDR1 transcript accumulation decreased in transgenic potato plants constitutively expressing a hairpin construct and these plants were challenged with three viruses: potato virus Y, potato virus X, and tobacco mosaic virus. Suppression of StRDR1 gene expression did not increase the susceptibility of potato to these viruses. Phylogenetic analysis of RDR genes present in potato and in a range of other plant species identified a new RDR gene family, not present in potato and found only in Rosids (but apparently lost in the Rosid A. thaliana) for which we propose the name RDR7.
Figures

Similar articles
-
RNA silencing-related genes contribute to tolerance of infection with potato virus X and Y in a susceptible tomato plant.Virol J. 2020 Oct 8;17(1):149. doi: 10.1186/s12985-020-01414-x. Virol J. 2020. PMID: 33032637 Free PMC article.
-
Salicylic acid treatment and expression of an RNA-dependent RNA polymerase 1 transgene inhibit lethal symptoms and meristem invasion during tobacco mosaic virus infection in Nicotiana benthamiana.BMC Plant Biol. 2016 Jan 13;16:15. doi: 10.1186/s12870-016-0705-8. BMC Plant Biol. 2016. PMID: 26757721 Free PMC article.
-
RNA-dependent RNA polymerase 1 from Nicotiana tabacum suppresses RNA silencing and enhances viral infection in Nicotiana benthamiana.Plant Cell. 2010 Apr;22(4):1358-72. doi: 10.1105/tpc.109.072058. Epub 2010 Apr 16. Plant Cell. 2010. PMID: 20400679 Free PMC article.
-
A review of host major-gene resistance to potato viruses X, Y, A and V in potato: genes, genetics and mapped locations.Heredity (Edinb). 2001 Jan;86(Pt 1):8-16. doi: 10.1046/j.1365-2540.2001.00798.x. Heredity (Edinb). 2001. PMID: 11298811 Review.
-
Breeding virus resistant potatoes (Solanum tuberosum): a review of traditional and molecular approaches.Heredity (Edinb). 2001 Jan;86(Pt 1):17-35. doi: 10.1046/j.1365-2540.2001.00799.x. Heredity (Edinb). 2001. PMID: 11298812 Review.
Cited by
-
Historical and Scientific Evidence for the Origin and Cultural Importance to Australia's First-Nations Peoples of the Laboratory Accession of Nicotiana benthamiana, a Model for Plant Virology.Viruses. 2022 Apr 8;14(4):771. doi: 10.3390/v14040771. Viruses. 2022. PMID: 35458501 Free PMC article.
-
Analysis of the RNA-Dependent RNA Polymerase 1 (RDR1) Gene Family in Melon.Plants (Basel). 2022 Jul 7;11(14):1795. doi: 10.3390/plants11141795. Plants (Basel). 2022. PMID: 35890429 Free PMC article.
-
Dynamics of a geminivirus-encoded pre-coat protein and host RNA-dependent RNA polymerase 1 in regulating symptom recovery in tobacco.J Exp Bot. 2018 Apr 9;69(8):2085-2102. doi: 10.1093/jxb/ery043. J Exp Bot. 2018. PMID: 29432546 Free PMC article.
-
CaRDR1, an RNA-Dependent RNA Polymerase Plays a Positive Role in Pepper Resistance against TMV.Front Plant Sci. 2017 Jun 28;8:1068. doi: 10.3389/fpls.2017.01068. eCollection 2017. Front Plant Sci. 2017. PMID: 28702034 Free PMC article.
-
Genome-wide identification and characterization of DCL, AGO and RDR gene families in Saccharum spontaneum.Sci Rep. 2020 Aug 6;10(1):13202. doi: 10.1038/s41598-020-70061-7. Sci Rep. 2020. PMID: 32764599 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

