A Brazilian Marseillevirus Is the Founding Member of a Lineage in Family Marseilleviridae
- PMID: 26978387
- PMCID: PMC4810266
- DOI: 10.3390/v8030076
A Brazilian Marseillevirus Is the Founding Member of a Lineage in Family Marseilleviridae
Abstract
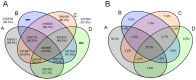
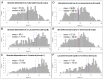
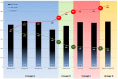
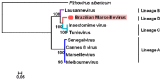
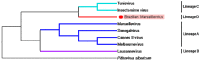
In 2003, Acanthamoeba polyphaga mimivirus (APMV) was discovered as parasitizing Acanthamoeba. It was revealed to exhibit remarkable features, especially odd genomic characteristics, and founded viral family Mimiviridae. Subsequently, a second family of giant amoebal viruses was described, Marseilleviridae, whose prototype member is Marseillevirus, discovered in 2009. Currently, the genomes of seven different members of this family have been fully sequenced. Previous phylogenetic analysis suggested the existence of three Marseilleviridae lineages: A, B and C. Here, we describe a new member of this family, Brazilian Marseillevirus (BrMV), which was isolated from a Brazilian sample and whose genome was fully sequenced and analyzed. Surprisingly, data from phylogenetic analyses and comparative genomics, including mean amino acid identity between BrMV and other Marseilleviridae members and the analyses of the core genome and pan-genome of marseilleviruses, indicated that this virus can be assigned to a new Marseilleviridae lineage. Even if the BrMV genome is one of the smallest among Marseilleviridae members, it harbors the second largest gene content into this family. In addition, the BrMV genome encodes 29 ORFans. Here, we describe the isolation and genome analyses of the BrMV strain, and propose its classification as the prototype virus of a new lineage D within the family Marseilleviridae.
Keywords: Brazilian marseillevirus; Marseilleviridae; Marseillevirus; genomic analyses; giant virus; lineage D.
Figures


Similar articles
-
Morphological and Taxonomic Properties of Tokyovirus, the First Marseilleviridae Member Isolated from Japan.Microbes Environ. 2016 Dec 23;31(4):442-448. doi: 10.1264/jsme2.ME16107. Epub 2016 Nov 19. Microbes Environ. 2016. PMID: 27867160 Free PMC article.
-
A new marseillevirus isolated in Southern Brazil from Limnoperna fortunei.Sci Rep. 2016 Oct 14;6:35237. doi: 10.1038/srep35237. Sci Rep. 2016. PMID: 27739526 Free PMC article.
-
The genomic and phylogenetic analysis of Marseillevirus cajuinensis raises questions about the evolution of Marseilleviridae lineages and their taxonomical organization.J Virol. 2024 Jun 13;98(6):e0051324. doi: 10.1128/jvi.00513-24. Epub 2024 May 16. J Virol. 2024. PMID: 38752754 Free PMC article.
-
The expanding family Marseilleviridae.Virology. 2014 Oct;466-467:27-37. doi: 10.1016/j.virol.2014.07.014. Epub 2014 Aug 5. Virology. 2014. PMID: 25104553 Review.
-
Gene repertoire of amoeba-associated giant viruses.Intervirology. 2010;53(5):330-43. doi: 10.1159/000312918. Epub 2010 Jun 15. Intervirology. 2010. PMID: 20551685 Review.
Cited by
-
Protozoal giant viruses: agents potentially infectious to humans and animals.Virus Genes. 2019 Oct;55(5):574-591. doi: 10.1007/s11262-019-01684-w. Epub 2019 Jul 9. Virus Genes. 2019. PMID: 31290063 Free PMC article. Review.
-
The Investigation of Promoter Sequences of Marseilleviruses Highlights a Remarkable Abundance of the AAATATTT Motif in Intergenic Regions.J Virol. 2017 Oct 13;91(21):e01088-17. doi: 10.1128/JVI.01088-17. Print 2017 Nov 1. J Virol. 2017. PMID: 28794030 Free PMC article.
-
A Brief History of Giant Viruses' Studies in Brazilian Biomes.Viruses. 2022 Jan 19;14(2):191. doi: 10.3390/v14020191. Viruses. 2022. PMID: 35215784 Free PMC article. Review.
-
Orpheovirus IHUMI-LCC2: A New Virus among the Giant Viruses.Front Microbiol. 2018 Jan 22;8:2643. doi: 10.3389/fmicb.2017.02643. eCollection 2017. Front Microbiol. 2018. PMID: 29403444 Free PMC article.
-
A Large Open Pangenome and a Small Core Genome for Giant Pandoraviruses.Front Microbiol. 2018 Jul 10;9:1486. doi: 10.3389/fmicb.2018.01486. eCollection 2018. Front Microbiol. 2018. PMID: 30042742 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

