Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives
- PMID: 26976576
- PMCID: PMC4822567
- DOI: 10.1073/pnas.1602472113
Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives
Abstract
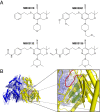
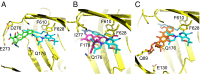
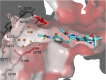
The Escherichia coli AcrAB-TolC efflux pump is the archetype of the resistance nodulation cell division (RND) exporters from Gram-negative bacteria. Overexpression of RND-type efflux pumps is a major factor in multidrug resistance (MDR), which makes these pumps important antibacterial drug discovery targets. We have recently developed novel pyranopyridine-based inhibitors of AcrB, which are orders of magnitude more powerful than the previously known inhibitors. However, further development of such inhibitors has been hindered by the lack of structural information for rational drug design. Although only the soluble, periplasmic part of AcrB binds and exports the ligands, the presence of the membrane-embedded domain in AcrB and its polyspecific binding behavior have made cocrystallization with drugs challenging. To overcome this obstacle, we have engineered and produced a soluble version of AcrB [AcrB periplasmic domain (AcrBper)], which is highly congruent in structure with the periplasmic part of the full-length protein, and is capable of binding substrates and potent inhibitors. Here, we describe the molecular basis for pyranopyridine-based inhibition of AcrB using a combination of cellular, X-ray crystallographic, and molecular dynamics (MD) simulations studies. The pyranopyridines bind within a phenylalanine-rich cage that branches from the deep binding pocket of AcrB, where they form extensive hydrophobic interactions. Moreover, the increasing potency of improved inhibitors correlates with the formation of a delicate protein- and water-mediated hydrogen bond network. These detailed insights provide a molecular platform for the development of novel combinational therapies using efflux pump inhibitors for combating multidrug resistant Gram-negative pathogens.
Keywords: RND efflux transporters; X-ray crystallography; efflux pump inhibitors; molecular dynamics simulation; multidrug resistance.
Conflict of interest statement
Conflict of interest statement: K.M.P. and reviewer B.L. coauthored a review article in 2015.
Figures
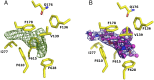


Similar articles
-
Molecular mechanism of MBX2319 inhibition of Escherichia coli AcrB multidrug efflux pump and comparison with other inhibitors.Antimicrob Agents Chemother. 2014 Oct;58(10):6224-34. doi: 10.1128/AAC.03283-14. Epub 2014 Aug 11. Antimicrob Agents Chemother. 2014. PMID: 25114133 Free PMC article.
-
Chlorpromazine and Amitriptyline Are Substrates and Inhibitors of the AcrB Multidrug Efflux Pump.mBio. 2020 Jun 2;11(3):e00465-20. doi: 10.1128/mBio.00465-20. mBio. 2020. PMID: 32487753 Free PMC article.
-
Kinetic analysis of the inhibition of the drug efflux protein AcrB using surface plasmon resonance.Biochim Biophys Acta Biomembr. 2018 Apr;1860(4):878-886. doi: 10.1016/j.bbamem.2017.08.024. Epub 2017 Sep 8. Biochim Biophys Acta Biomembr. 2018. PMID: 28890187
-
AcrB: a mean, keen, drug efflux machine.Ann N Y Acad Sci. 2020 Jan;1459(1):38-68. doi: 10.1111/nyas.14239. Epub 2019 Oct 6. Ann N Y Acad Sci. 2020. PMID: 31588569 Review.
-
Optimization of a novel series of pyranopyridine RND efflux pump inhibitors.Curr Opin Microbiol. 2016 Oct;33:1-6. doi: 10.1016/j.mib.2016.05.007. Epub 2016 May 24. Curr Opin Microbiol. 2016. PMID: 27232955 Free PMC article. Review.
Cited by
-
Identification of a Novel Polyamine Scaffold With Potent Efflux Pump Inhibition Activity Toward Multi-Drug Resistant Bacterial Pathogens.Front Microbiol. 2018 Jun 14;9:1301. doi: 10.3389/fmicb.2018.01301. eCollection 2018. Front Microbiol. 2018. PMID: 29963035 Free PMC article.
-
Mapping the Dynamic Functions and Structural Features of AcrB Efflux Pump Transporter Using Accelerated Molecular Dynamics Simulations.Sci Rep. 2018 Jul 11;8(1):10470. doi: 10.1038/s41598-018-28531-6. Sci Rep. 2018. PMID: 29992991 Free PMC article.
-
Tripartite efflux pumps of the RND superfamily: what did we learn from computational studies?Microbiology (Reading). 2023 Mar;169(3):001307. doi: 10.1099/mic.0.001307. Microbiology (Reading). 2023. PMID: 36972322 Free PMC article. Review.
-
An efflux-susceptible antibiotic-adjuvant with systemic efficacy against mouse infections.Sci Rep. 2022 Oct 21;12(1):17673. doi: 10.1038/s41598-022-21526-4. Sci Rep. 2022. PMID: 36271103 Free PMC article.
-
Microbial Efflux Pump Inhibitors: A Journey around Quinoline and Indole Derivatives.Molecules. 2021 Nov 19;26(22):6996. doi: 10.3390/molecules26226996. Molecules. 2021. PMID: 34834098 Free PMC article. Review.
References
-
- Ruggerone P, Murakami S, Pos KM, Vargiu AV. RND efflux pumps: Structural information translated into function and inhibition mechanisms. Curr Top Med Chem. 2013;13(24):3079–3100. - PubMed
-
- Du D, van Veen HW, Murakami S, Pos KM, Luisi BF. Structure, mechanism and cooperation of bacterial multidrug transporters. Curr Opin Struct Biol. 2015;33:76–91. - PubMed
-
- Murakami S, Nakashima R, Yamashita E, Matsumoto T, Yamaguchi A. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature. 2006;443(7108):173–179. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

