Display of fungal hydrophobin on the Pichia pastoris cell surface and its influence on Candida antarctica lipase B
- PMID: 26969039
- PMCID: PMC4911288
- DOI: 10.1007/s00253-016-7431-x
Display of fungal hydrophobin on the Pichia pastoris cell surface and its influence on Candida antarctica lipase B
Abstract
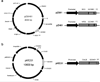
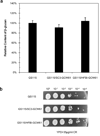
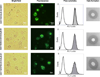
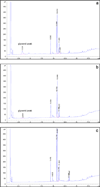
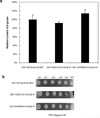
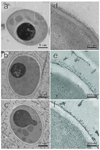
To modify the Pichia pastoris cell surface, two classes of hydrophobins, SC3 from Schizophyllum commune and HFBI from Trichoderma reesei, were separately displayed on the cell wall. There was an observable increase in the hydrophobicity of recombinant strains. Candida antarctica lipase B (CALB) was then co-displayed on the modified cells, generating strains GS115/SC3-61/CALB-51 and GS115/HFBI-61/CALB-51. Interestingly, the hydrolytic and synthetic activities of strain GS115/HFBI-61/CALB-51 increased by 37 and 109 %, respectively, but decreased by 26 and 43 %, respectively, in strain GS115/SC3-61/CALB-51 compared with the hydrophobin-minus recombinant strain GS115/CALB-GCW51. The amount of glycerol by-product from the transesterification reaction adsorbed on the cell surface was significantly decreased following hydrophobin modification, removing the glycerol barrier and allowing substrates to access the active sites of lipases. Electron micrographs indicated that the cell wall structures of both recombinant strains appeared altered, including changes to the inner glucan layer and outer mannan layer. These results suggest that the display of hydrophobins can change the surface structure and hydrophobic properties of P. pastoris and affect the catalytic activities of CALB displayed on the surface of P. pastoris cells.
Keywords: Candida antarctica lipase B; Co-display; Hydrophobicity; Hydrophobin; Pichia pastoris.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
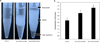



Similar articles
-
Accurate analysis of fusion expression of Pichia pastoris glycosylphosphatidylinositol-modified cell wall proteins.J Ind Microbiol Biotechnol. 2017 Sep;44(9):1355-1365. doi: 10.1007/s10295-017-1962-8. Epub 2017 Jun 28. J Ind Microbiol Biotechnol. 2017. PMID: 28660369
-
Display of Candida antarctica lipase B on Pichia pastoris and its application to flavor ester synthesis.Appl Microbiol Biotechnol. 2010 May;86(5):1493-501. doi: 10.1007/s00253-009-2382-0. Epub 2009 Dec 24. Appl Microbiol Biotechnol. 2010. PMID: 20033404
-
Double Candida antarctica lipase B co-display on Pichia pastoris cell surface based on a self-processing foot-and-mouth disease virus 2A peptide.Appl Microbiol Biotechnol. 2012 Dec;96(6):1539-50. doi: 10.1007/s00253-012-4264-0. Epub 2012 Jul 14. Appl Microbiol Biotechnol. 2012. PMID: 22797600
-
Recent Advances in Pichia pastoris as Host for Heterologous Expression System for Lipases: A Review.Methods Mol Biol. 2018;1835:205-216. doi: 10.1007/978-1-4939-8672-9_11. Methods Mol Biol. 2018. PMID: 30109654 Review.
-
Recent Advances in Fungal Hydrophobin Towards Using in Industry.Protein J. 2015 Aug;34(4):243-55. doi: 10.1007/s10930-015-9621-2. Protein J. 2015. PMID: 26208665 Review.
Cited by
-
Accurate analysis of fusion expression of Pichia pastoris glycosylphosphatidylinositol-modified cell wall proteins.J Ind Microbiol Biotechnol. 2017 Sep;44(9):1355-1365. doi: 10.1007/s10295-017-1962-8. Epub 2017 Jun 28. J Ind Microbiol Biotechnol. 2017. PMID: 28660369
-
Effects of Enzyme Loading and Immobilization Conditions on the Catalytic Features of Lipase From Pseudomonas fluorescens Immobilized on Octyl-Agarose Beads.Front Bioeng Biotechnol. 2020 Feb 28;8:36. doi: 10.3389/fbioe.2020.00036. eCollection 2020. Front Bioeng Biotechnol. 2020. PMID: 32181245 Free PMC article.
-
Cryo-FIB specimen preparation for use in a cartridge-type cryo-TEM.J Struct Biol. 2017 Aug;199(2):114-119. doi: 10.1016/j.jsb.2017.05.011. Epub 2017 May 27. J Struct Biol. 2017. PMID: 28559166 Free PMC article.
-
Surface display of HFBI and DewA hydrophobins on Saccharomyces cerevisiae modifies tolerance to several adverse conditions and biocatalytic performance.Appl Microbiol Biotechnol. 2021 Feb;105(4):1505-1518. doi: 10.1007/s00253-021-11090-8. Epub 2021 Jan 23. Appl Microbiol Biotechnol. 2021. PMID: 33484321
-
Role of Hydrophobins in Aspergillus fumigatus.J Fungi (Basel). 2017 Dec 24;4(1):2. doi: 10.3390/jof4010002. J Fungi (Basel). 2017. PMID: 29371496 Free PMC article.
References
-
- Bastida A, Sabuquillo P, Armisen P, Fernandez-Lafuente R, Huguet J, Guisan JM. A single step purification, immobilization, and hyperactivation of lipases via interfacial adsorption on strongly hydrophobic supports. Biotechnol Bioeng. 1998;58(5):486–493. - PubMed
-
- Brzozowski A, Derewenda U, Derewenda Z, Dodson G, Lawson D, Turkenburg J, Bjorkling F, Huge-Jensen B, Patkar S, Thim L. A model for interfacial activation in lipases from the structure of a fungal lipase-inhibitor complex. Nature. 1991;351(6326):491–494. - PubMed
-
- Chen B, Pernodet N, Rafailovich MH, Bakhtina A, Gross RA. Protein immobilization on epoxy-activated thin polymer films: effect of surface wettability and enzyme loading. Langmuir. 2008;24(23):13457–13464. - PubMed
-
- Espino-Rammer L, Ribitsch D, Przylucka A, Marold A, Greimel KJ, Acero EH, Guebitz GM, Kubicek CP, Druzhinina IS. Two novel class II hydrophobins from Trichoderma spp. stimulate enzymatic hydrolysis of poly (ethylene terephthalate) when expressed as fusion proteins. Appl Environ Microb. 2013;79(14):4230–4238. - PMC - PubMed
-
- Hama S, Yoshida A, Nakashima K, Noda H, Fukuda H, Kondo A. Surfactant-modified yeast whole-cell biocatalyst displaying lipase on cell surface for enzymatic production of structured lipids in organic media. Appl Microbiol Biot. 2010;87(2):537–543. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

