Efficient and biologically relevant consensus strategy for Parkinson's disease gene prioritization
- PMID: 26961748
- PMCID: PMC4784386
- DOI: 10.1186/s12920-016-0173-x
Efficient and biologically relevant consensus strategy for Parkinson's disease gene prioritization
Abstract
Background: The systemic information enclosed in microarray data encodes relevant clues to overcome the poorly understood combination of genetic and environmental factors in Parkinson's disease (PD), which represents the major obstacle to understand its pathogenesis and to develop disease-modifying therapeutics. While several gene prioritization approaches have been proposed, none dominate over the rest. Instead, hybrid approaches seem to outperform individual approaches.
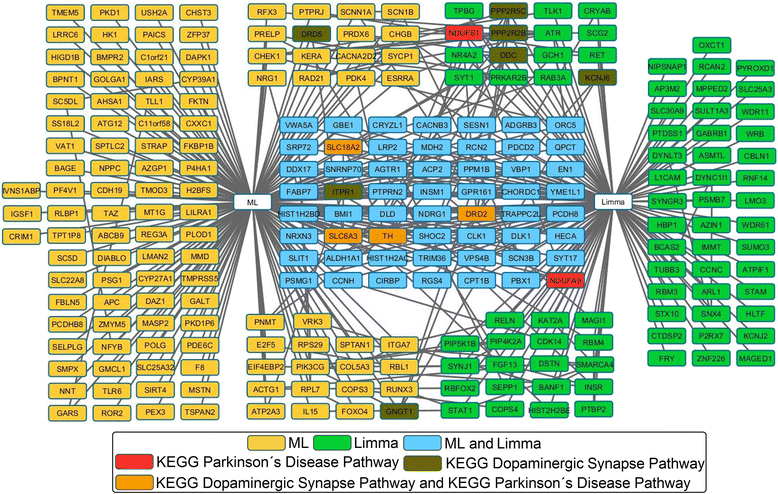
Methods: A consensus strategy is proposed for PD related gene prioritization from mRNA microarray data based on the combination of three independent prioritization approaches: Limma, machine learning, and weighted gene co-expression networks.
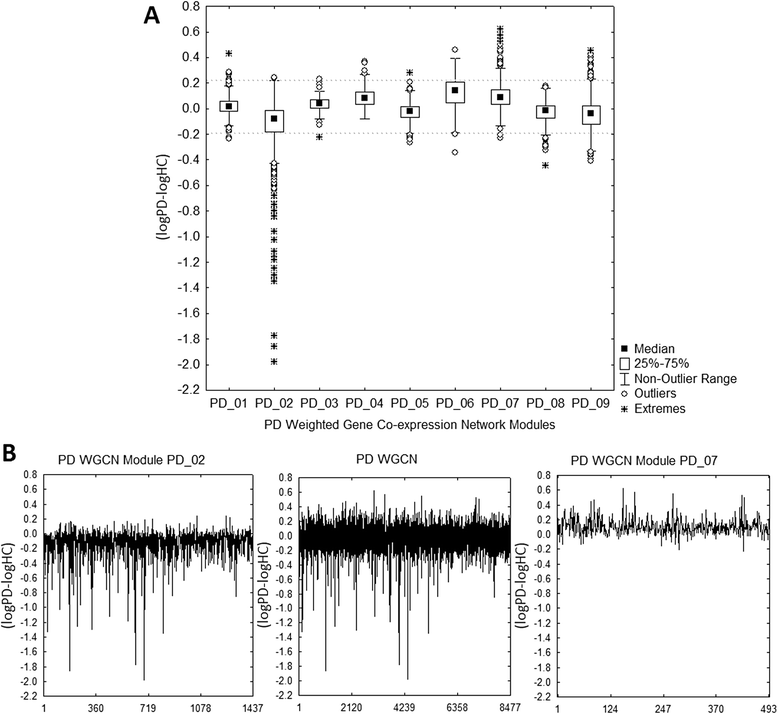
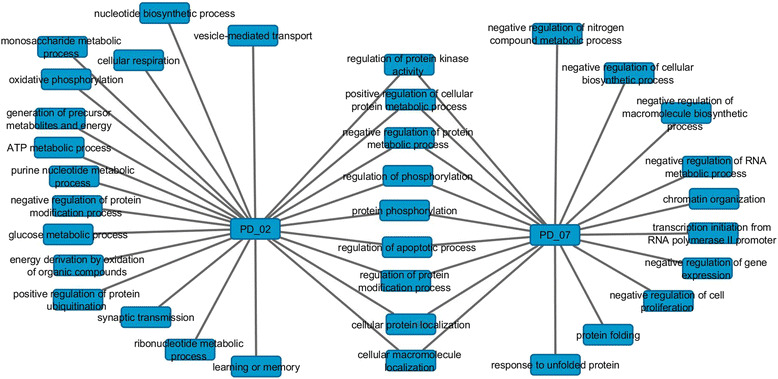
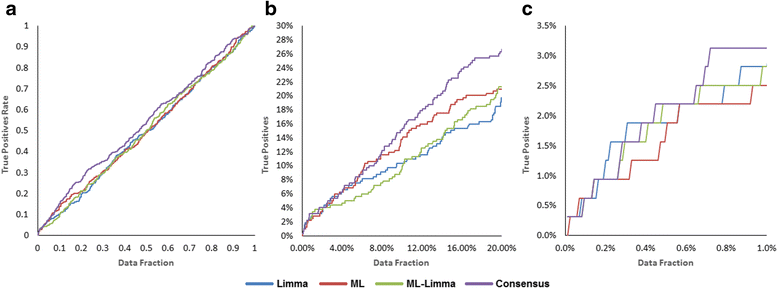
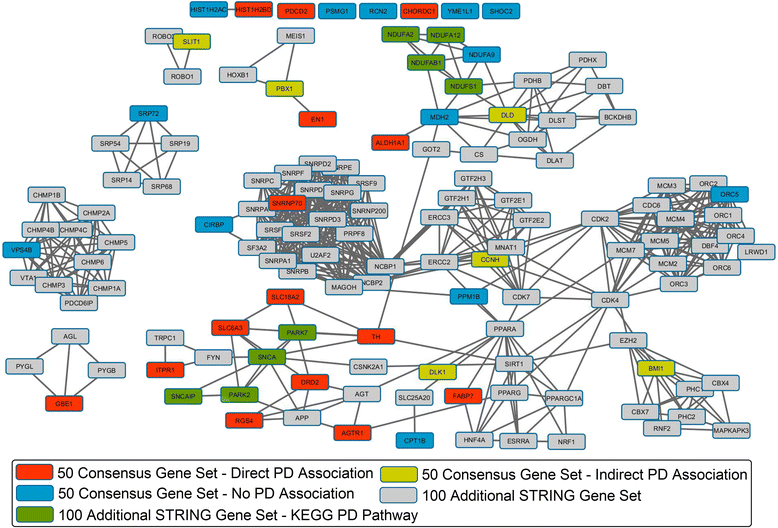
Results: The consensus strategy outperformed the individual approaches in terms of statistical significance, overall enrichment and early recognition ability. In addition to a significant biological relevance, the set of 50 genes prioritized exhibited an excellent early recognition ability (6 of the top 10 genes are directly associated with PD). 40 % of the prioritized genes were previously associated with PD including well-known PD related genes such as SLC18A2, TH or DRD2. Eight genes (CCNH, DLK1, PCDH8, SLIT1, DLD, PBX1, INSM1, and BMI1) were found to be significantly associated to biological process affected in PD, representing potentially novel PD biomarkers or therapeutic targets. Additionally, several metrics of standard use in chemoinformatics are proposed to evaluate the early recognition ability of gene prioritization tools.
Conclusions: The proposed consensus strategy represents an efficient and biologically relevant approach for gene prioritization tasks providing a valuable decision-making tool for the study of PD pathogenesis and the development of disease-modifying PD therapeutics.
Figures
Similar articles
-
Identification of potential immune-related hub genes in Parkinson's disease based on machine learning and development and validation of a diagnostic classification model.PLoS One. 2023 Dec 5;18(12):e0294984. doi: 10.1371/journal.pone.0294984. eCollection 2023. PLoS One. 2023. PMID: 38051734 Free PMC article.
-
Construction of Parkinson's disease marker-based weighted protein-protein interaction network for prioritization of co-expressed genes.Gene. 2019 May 20;697:67-77. doi: 10.1016/j.gene.2019.02.026. Epub 2019 Feb 16. Gene. 2019. PMID: 30776463
-
Repositioning drugs by targeting network modules: a Parkinson's disease case study.BMC Bioinformatics. 2017 Dec 28;18(Suppl 14):532. doi: 10.1186/s12859-017-1889-0. BMC Bioinformatics. 2017. PMID: 29297292 Free PMC article.
-
Contribution of genomics and proteomics in understanding the role of modifying factors in Parkinson's disease.Indian J Biochem Biophys. 2006 Apr;43(2):69-81. Indian J Biochem Biophys. 2006. PMID: 16955754 Review.
-
Genetic risk factors in Parkinson's disease.Cell Tissue Res. 2018 Jul;373(1):9-20. doi: 10.1007/s00441-018-2817-y. Epub 2018 Mar 13. Cell Tissue Res. 2018. PMID: 29536161 Free PMC article. Review.
Cited by
-
Gene prioritization, communality analysis, networking and metabolic integrated pathway to better understand breast cancer pathogenesis.Sci Rep. 2018 Nov 12;8(1):16679. doi: 10.1038/s41598-018-35149-1. Sci Rep. 2018. PMID: 30420728 Free PMC article.
-
Alzheimer disease: A quantitative trait approach to GWAS pays dividends.Nat Rev Neurol. 2017 Jun;13(6):321-322. doi: 10.1038/nrneurol.2017.61. Epub 2017 Apr 28. Nat Rev Neurol. 2017. PMID: 28452353 No abstract available.
-
Concordance analysis of microarray studies identifies representative gene expression changes in Parkinson's disease: a comparison of 33 human and animal studies.BMC Neurol. 2017 Mar 23;17(1):58. doi: 10.1186/s12883-017-0838-x. BMC Neurol. 2017. PMID: 28335819 Free PMC article.
-
Comparison of multiple transcriptomes exposes unified and divergent features of quiescent and activated skeletal muscle stem cells.Skelet Muscle. 2017 Dec 22;7(1):28. doi: 10.1186/s13395-017-0144-8. Skelet Muscle. 2017. PMID: 29273087 Free PMC article.
-
Meta-Analysis of Differentially Expressed Genes in the Substantia Nigra in Parkinson's Disease Supports Phenotype-Specific Transcriptome Changes.Front Neurosci. 2020 Dec 18;14:596105. doi: 10.3389/fnins.2020.596105. eCollection 2020. Front Neurosci. 2020. PMID: 33390883 Free PMC article.
References
-
- de Lau LM, Giesbergen PC, de Rijk MC, Hofman A, Koudstaal PJ, Breteler MM. Incidence of parkinsonism and Parkinson disease in a general population: the Rotterdam Study. Neurology. 2004;63(7):1240–4. doi: 10.1212/01.WNL.0000140706.52798.BE. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

