Acetylation mimic of lysine 280 exacerbates human Tau neurotoxicity in vivo
- PMID: 26940749
- PMCID: PMC4778021
- DOI: 10.1038/srep22685
Acetylation mimic of lysine 280 exacerbates human Tau neurotoxicity in vivo
Abstract
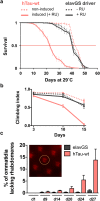
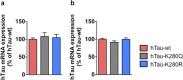
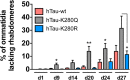
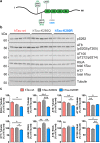
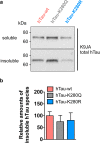
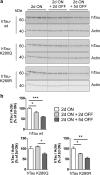
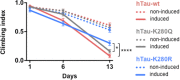
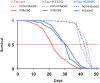
Dysfunction and accumulation of the microtubule-associated human Tau (hTau) protein into intraneuronal aggregates is observed in many neurodegenerative disorders including Alzheimer's disease (AD). Reversible lysine acetylation has recently emerged as a post-translational modification that may play an important role in the modulation of hTau pathology. Acetylated hTau species have been observed within hTau aggregates in human AD brains and multi-acetylation of hTau in vitro regulates its propensity to aggregate. However, whether lysine acetylation at position 280 (K280) modulates hTau-induced toxicity in vivo is unknown. We generated new Drosophila transgenic models of hTau pathology to evaluate the contribution of K280 acetylation to hTau toxicity, by analysing the respective toxicity of pseudo-acetylated (K280Q) and pseudo-de-acetylated (K280R) mutant forms of hTau. We observed that mis-expression of pseudo-acetylated K280Q-hTau in the adult fly nervous system potently exacerbated fly locomotion defects and photoreceptor neurodegeneration. In addition, modulation of K280 influenced total hTau levels and phosphorylation without changing hTau solubility. Altogether, our results indicate that pseudo-acetylation of the single K280 residue is sufficient to exacerbate hTau neurotoxicity in vivo, suggesting that acetylated K280-hTau species contribute to the pathological events leading to neurodegeneration in AD.
Figures
Similar articles
-
Pseudo-acetylation of multiple sites on human Tau proteins alters Tau phosphorylation and microtubule binding, and ameliorates amyloid beta toxicity.Sci Rep. 2017 Aug 30;7(1):9984. doi: 10.1038/s41598-017-10225-0. Sci Rep. 2017. PMID: 28855586 Free PMC article.
-
The acetylation of tau inhibits its function and promotes pathological tau aggregation.Nat Commun. 2011;2:252. doi: 10.1038/ncomms1255. Nat Commun. 2011. PMID: 21427723 Free PMC article.
-
A unique tau conformation generated by an acetylation-mimic substitution modulates P301S-dependent tau pathology and hyperphosphorylation.J Biol Chem. 2019 Nov 8;294(45):16698-16711. doi: 10.1074/jbc.RA119.009674. Epub 2019 Sep 22. J Biol Chem. 2019. PMID: 31543505 Free PMC article.
-
Cellular and molecular modifier pathways in tauopathies: the big picture from screening invertebrate models.J Neurochem. 2016 Apr;137(1):12-25. doi: 10.1111/jnc.13532. Epub 2016 Feb 11. J Neurochem. 2016. PMID: 26756400 Review.
-
Regulation of Tau Homeostasis and Toxicity by Acetylation.Adv Exp Med Biol. 2019;1184:47-55. doi: 10.1007/978-981-32-9358-8_4. Adv Exp Med Biol. 2019. PMID: 32096027 Review.
Cited by
-
Uncoupling neuronal death and dysfunction in Drosophila models of neurodegenerative disease.Acta Neuropathol Commun. 2016 Jun 23;4(1):62. doi: 10.1186/s40478-016-0333-4. Acta Neuropathol Commun. 2016. PMID: 27338814 Free PMC article.
-
A hybrid approach unveils drug repurposing candidates targeting an Alzheimer pathophysiology mechanism.Patterns (N Y). 2022 Jan 26;3(3):100433. doi: 10.1016/j.patter.2021.100433. eCollection 2022 Mar 11. Patterns (N Y). 2022. PMID: 35510183 Free PMC article.
-
Regulation of INF2-mediated actin polymerization through site-specific lysine acetylation of actin itself.Proc Natl Acad Sci U S A. 2020 Jan 7;117(1):439-447. doi: 10.1073/pnas.1914072117. Epub 2019 Dec 23. Proc Natl Acad Sci U S A. 2020. PMID: 31871199 Free PMC article.
-
Degradation or aggregation: the ramifications of post-translational modifications on tau.BMB Rep. 2018 Jun;51(6):265-273. doi: 10.5483/bmbrep.2018.51.6.077. BMB Rep. 2018. PMID: 29661268 Free PMC article. Review.
-
Post-Translational Modifications in Tau and Their Roles in Alzheimer's Pathology.Curr Alzheimer Res. 2024;21(1):24-49. doi: 10.2174/0115672050301407240408033046. Curr Alzheimer Res. 2024. PMID: 38623984 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

