Conformational Flexibility Enables the Function of a BECN1 Region Essential for Starvation-Mediated Autophagy
- PMID: 26937551
- PMCID: PMC4876825
- DOI: 10.1021/acs.biochem.5b01264
Conformational Flexibility Enables the Function of a BECN1 Region Essential for Starvation-Mediated Autophagy
Abstract
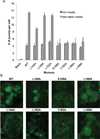
BECN1 is essential for autophagy, a critical eukaryotic cellular homeostasis pathway. Here we delineate a highly conserved BECN1 domain located between previously characterized BH3 and coiled-coil domains and elucidate its structure and role in autophagy. The 2.0 Å sulfur-single-wavelength anomalous dispersion X-ray crystal structure of this domain demonstrates that its N-terminal half is unstructured while its C-terminal half is helical; hence, we name it the flexible helical domain (FHD). Circular dichroism spectroscopy, double electron-electron resonance-electron paramagnetic resonance, and small-angle X-ray scattering (SAXS) analyses confirm that the FHD is partially disordered, even in the context of adjacent BECN1 domains. Molecular dynamic simulations fitted to SAXS data indicate that the FHD transiently samples more helical conformations. FHD helicity increases in 2,2,2-trifluoroethanol, suggesting it may become more helical upon binding. Lastly, cellular studies show that conserved FHD residues are required for starvation-induced autophagy. Thus, the FHD likely undergoes a binding-associated disorder-to-helix transition, and conserved residues critical for this interaction are essential for starvation-induced autophagy.
Figures


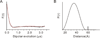




Similar articles
-
Structural transitions in conserved, ordered Beclin 1 domains essential to regulating autophagy.J Biol Chem. 2017 Sep 29;292(39):16235-16248. doi: 10.1074/jbc.M117.804195. Epub 2017 Aug 10. J Biol Chem. 2017. PMID: 28798234 Free PMC article.
-
Conformational flexibility of BECN1: Essential to its key role in autophagy and beyond.Protein Sci. 2016 Oct;25(10):1767-85. doi: 10.1002/pro.2984. Epub 2016 Aug 13. Protein Sci. 2016. PMID: 27414988 Free PMC article. Review.
-
Identification of BECN1 and ATG14 Coiled-Coil Interface Residues That Are Important for Starvation-Induced Autophagy.Biochemistry. 2016 Aug 2;55(30):4239-53. doi: 10.1021/acs.biochem.6b00246. Epub 2016 Jul 22. Biochemistry. 2016. PMID: 27383850 Free PMC article.
-
Intrinsically disordered regions in autophagy proteins.Proteins. 2014 Apr;82(4):565-78. doi: 10.1002/prot.24424. Epub 2013 Oct 17. Proteins. 2014. PMID: 24115198 Free PMC article.
-
The autophagy effector Beclin 1: a novel BH3-only protein.Oncogene. 2008 Dec;27 Suppl 1(Suppl 1):S137-48. doi: 10.1038/onc.2009.51. Oncogene. 2008. PMID: 19641499 Free PMC article. Review.
Cited by
-
Autophagy-Related Beclin 1 and Head and Neck Cancers.Onco Targets Ther. 2020 Jun 30;13:6213-6227. doi: 10.2147/OTT.S256072. eCollection 2020. Onco Targets Ther. 2020. PMID: 32669852 Free PMC article. Review.
-
Structural transitions in conserved, ordered Beclin 1 domains essential to regulating autophagy.J Biol Chem. 2017 Sep 29;292(39):16235-16248. doi: 10.1074/jbc.M117.804195. Epub 2017 Aug 10. J Biol Chem. 2017. PMID: 28798234 Free PMC article.
-
Targeting autophagy and beyond: Deconvoluting the complexity of Beclin-1 from biological function to cancer therapy.Acta Pharm Sin B. 2023 Dec;13(12):4688-4714. doi: 10.1016/j.apsb.2023.08.008. Epub 2023 Aug 12. Acta Pharm Sin B. 2023. PMID: 38045051 Free PMC article. Review.
-
Conformational flexibility of BECN1: Essential to its key role in autophagy and beyond.Protein Sci. 2016 Oct;25(10):1767-85. doi: 10.1002/pro.2984. Epub 2016 Aug 13. Protein Sci. 2016. PMID: 27414988 Free PMC article. Review.
-
Vps34 Kinase Domain Dynamics Regulate the Autophagic PI 3-Kinase Complex.Mol Cell. 2017 Aug 3;67(3):528-534.e3. doi: 10.1016/j.molcel.2017.07.003. Epub 2017 Jul 27. Mol Cell. 2017. PMID: 28757208 Free PMC article.
References
-
- Levine B, Klionsky DJ. Development by self-digestion: Molecular mechanisms and biological functions of autophagy. Dev. Cell. 2004;6:463–477. - PubMed
-
- Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9 - PubMed
-
- Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 2011;27:107–132. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

