Complex Interplay of the UL136 Isoforms Balances Cytomegalovirus Replication and Latency
- PMID: 26933055
- PMCID: PMC4810493
- DOI: 10.1128/mBio.01986-15
Complex Interplay of the UL136 Isoforms Balances Cytomegalovirus Replication and Latency
Abstract
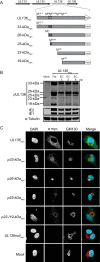
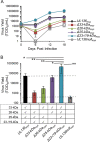
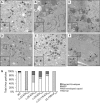
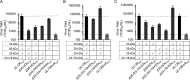
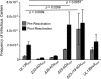
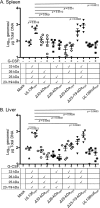
Human cytomegalovirus (HCMV), a betaherpesvirus, persists indefinitely in the human host through poorly understood mechanisms. The UL136 gene is carried within a genetic locus important to HCMV latency termed the UL133/8 locus, which also carries UL133, UL135, and UL138. Previously, we demonstrated that UL136 is expressed as five protein isoforms ranging from 33-kDa to 19-kDa, arising from alternative transcription and, likely, translation initiation mechanisms. We previously showed that the UL136 isoforms are largely dispensable for virus infection in fibroblasts, a model for productive virus replication. In our current work, UL136 has emerged as a complex regulator of HCMV infection in multiple contexts of infection relevant to HCMV persistence: in an endothelial cell (EC) model of chronic infection, in a CD34(+) hematopoietic progenitor cell (HPC) model of latency, and in an in vivo NOD-scid IL2Rγc (null) humanized (huNSG) mouse model for latency. The 33- and 26-kDa isoforms promote replication, while the 23- and 19-kDa isoforms suppress replication in ECs, in CD34(+) HPCs, and in huNSG mice. The role of the 25-kDa isoform is context dependent and influences the activity of the other isoforms. These isoforms localize throughout the secretory pathway, and loss of the 33- and 26-kDa UL136 isoforms results in virus maturation defects in ECs. This work reveals an intriguing functional interplay between protein isoforms that impacts virus replication, latency, and dissemination, contributing to the overall role of the UL133/8 locus in HCMV infection.
Importance: The persistence of DNA viruses, and particularly of herpesviruses, remains an enigma because we have not completely defined the viral and host factors important to persistence. Human cytomegalovirus, a herpesvirus, persists in the absence of disease in immunocompetent individuals but poses a serious disease threat to transplant patients and the developing fetus. There is no vaccine, and current therapies do not target latent reservoirs. In an effort to define the viral factors important to persistence, we have studied viral genes with no known viral replication function in contexts important to HCMV persistence. Using models relevant to viral persistence, we demonstrate opposing roles of protein isoforms encoded by the UL136 gene in regulating latent and replicative states of infection. Our findings reveal an intriguing interplay between UL136 protein isoforms and define UL136 as an important regulator of HCMV persistence.
Copyright © 2016 Caviness et al.
Figures


Similar articles
-
Complex expression of the UL136 gene of human cytomegalovirus results in multiple protein isoforms with unique roles in replication.J Virol. 2014 Dec;88(24):14412-25. doi: 10.1128/JVI.02711-14. Epub 2014 Oct 8. J Virol. 2014. PMID: 25297993 Free PMC article.
-
Stabilization of the human cytomegalovirus UL136p33 reactivation determinant overcomes the requirement for UL135 for replication in hematopoietic cells.J Virol. 2023 Aug 31;97(8):e0014823. doi: 10.1128/jvi.00148-23. Epub 2023 Aug 11. J Virol. 2023. PMID: 37565749 Free PMC article.
-
Human Cytomegalovirus UL135 and UL136 Genes Are Required for Postentry Tropism in Endothelial Cells.J Virol. 2015 Jul;89(13):6536-50. doi: 10.1128/JVI.00284-15. Epub 2015 Apr 15. J Virol. 2015. PMID: 25878111 Free PMC article.
-
The Role of the Human Cytomegalovirus UL133-UL138 Gene Locus in Latency and Reactivation.Viruses. 2020 Jul 1;12(7):714. doi: 10.3390/v12070714. Viruses. 2020. PMID: 32630219 Free PMC article. Review.
-
Human cytomegalovirus persistence.Cell Microbiol. 2012 May;14(5):644-55. doi: 10.1111/j.1462-5822.2012.01774.x. Epub 2012 Mar 8. Cell Microbiol. 2012. PMID: 22329758 Free PMC article. Review.
Cited by
-
Advances in Model Systems for Human Cytomegalovirus Latency and Reactivation.mBio. 2022 Feb 22;13(1):e0172421. doi: 10.1128/mbio.01724-21. Epub 2022 Jan 11. mBio. 2022. PMID: 35012351 Free PMC article. Review.
-
Control of Immediate Early Gene Expression for Human Cytomegalovirus Reactivation.Front Cell Infect Microbiol. 2020 Sep 17;10:476. doi: 10.3389/fcimb.2020.00476. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 33072616 Free PMC article. Review.
-
HCMV miR-US22 down-regulation of EGR-1 regulates CD34+ hematopoietic progenitor cell proliferation and viral reactivation.PLoS Pathog. 2019 Nov 14;15(11):e1007854. doi: 10.1371/journal.ppat.1007854. eCollection 2019 Nov. PLoS Pathog. 2019. PMID: 31725809 Free PMC article.
-
HCMV miRNA Targets Reveal Important Cellular Pathways for Viral Replication, Latency, and Reactivation.Noncoding RNA. 2018 Oct 22;4(4):29. doi: 10.3390/ncrna4040029. Noncoding RNA. 2018. PMID: 30360396 Free PMC article. Review.
-
Molecular Determinants and the Regulation of Human Cytomegalovirus Latency and Reactivation.Viruses. 2018 Aug 20;10(8):444. doi: 10.3390/v10080444. Viruses. 2018. PMID: 30127257 Free PMC article. Review.
References
-
- Mocarski ES, Shenk TE, Griffiths PD, Pass RF. 2013. Cytomegaloviruses. In Fields’ virology, 6th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
-
- Wang GC, Kao WH, Murakami P, Xue Q-L, Chiou RB, Detrick B, McDyer JF, Semba RD, Casolaro V, Walston JD, Fried LP. 2010. Cytomegalovirus infection and the risk of mortality and frailty in older women: a prospective observational cohort study. Am J Epidemiol 171:1144–1152. doi:10.1093/aje/kwq062. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
