ALIX and ESCRT-I/II function as parallel ESCRT-III recruiters in cytokinetic abscission
- PMID: 26929449
- PMCID: PMC4772496
- DOI: 10.1083/jcb.201507009
ALIX and ESCRT-I/II function as parallel ESCRT-III recruiters in cytokinetic abscission
Abstract
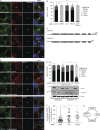
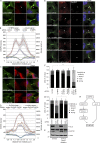
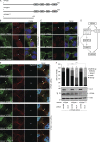
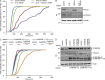
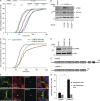
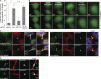
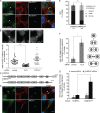
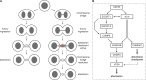
Cytokinetic abscission, the final stage of cell division where the two daughter cells are separated, is mediated by the endosomal sorting complex required for transport (ESCRT) machinery. The ESCRT-III subunit CHMP4B is a key effector in abscission, whereas its paralogue, CHMP4C, is a component in the abscission checkpoint that delays abscission until chromatin is cleared from the intercellular bridge. How recruitment of these components is mediated during cytokinesis remains poorly understood, although the ESCRT-binding protein ALIX has been implicated. Here, we show that ESCRT-II and the ESCRT-II-binding ESCRT-III subunit CHMP6 cooperate with ESCRT-I to recruit CHMP4B, with ALIX providing a parallel recruitment arm. In contrast to CHMP4B, we find that recruitment of CHMP4C relies predominantly on ALIX. Accordingly, ALIX depletion leads to furrow regression in cells with chromosome bridges, a phenotype associated with abscission checkpoint signaling failure. Collectively, our work reveals a two-pronged recruitment of ESCRT-III to the cytokinetic bridge and implicates ALIX in abscission checkpoint signaling.
© 2016 Christ et al.
Figures
Comment in
-
Burning cellular bridges: Two pathways to the big breakup.J Cell Biol. 2016 Feb 29;212(5):491-3. doi: 10.1083/jcb.201602003. J Cell Biol. 2016. PMID: 26929447 Free PMC article.
Similar articles
-
The Flemmingsome reveals an ESCRT-to-membrane coupling via ALIX/syntenin/syndecan-4 required for completion of cytokinesis.Nat Commun. 2020 Apr 22;11(1):1941. doi: 10.1038/s41467-020-15205-z. Nat Commun. 2020. PMID: 32321914 Free PMC article.
-
Vesicle-mediated transport of ALIX and ESCRT-III to the intercellular bridge during cytokinesis.Cell Mol Life Sci. 2023 Jul 31;80(8):235. doi: 10.1007/s00018-023-04864-y. Cell Mol Life Sci. 2023. PMID: 37523003 Free PMC article.
-
A Septin Double Ring Controls the Spatiotemporal Organization of the ESCRT Machinery in Cytokinetic Abscission.Curr Biol. 2019 Jul 8;29(13):2174-2182.e7. doi: 10.1016/j.cub.2019.05.050. Epub 2019 Jun 13. Curr Biol. 2019. PMID: 31204162 Free PMC article.
-
The Abscission Checkpoint: Making It to the Final Cut.Trends Cell Biol. 2017 Jan;27(1):1-11. doi: 10.1016/j.tcb.2016.10.001. Epub 2016 Oct 31. Trends Cell Biol. 2017. PMID: 27810282 Review.
-
Knowing when to cut and run: mechanisms that control cytokinetic abscission.Trends Cell Biol. 2013 Sep;23(9):433-41. doi: 10.1016/j.tcb.2013.04.006. Epub 2013 May 22. Trends Cell Biol. 2013. PMID: 23706391 Review.
Cited by
-
A cancer-associated polymorphism in ESCRT-III disrupts the abscission checkpoint and promotes genome instability.Proc Natl Acad Sci U S A. 2018 Sep 18;115(38):E8900-E8908. doi: 10.1073/pnas.1805504115. Epub 2018 Sep 4. Proc Natl Acad Sci U S A. 2018. PMID: 30181294 Free PMC article.
-
Contribution of integrin adhesion to cytokinetic abscission and genomic integrity.Front Cell Dev Biol. 2022 Dec 12;10:1048717. doi: 10.3389/fcell.2022.1048717. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36578785 Free PMC article. Review.
-
PI(3,4)P2-mediated cytokinetic abscission prevents early senescence and cataract formation.Science. 2021 Dec 10;374(6573):eabk0410. doi: 10.1126/science.abk0410. Epub 2021 Dec 10. Science. 2021. PMID: 34882480 Free PMC article.
-
The Flemmingsome reveals an ESCRT-to-membrane coupling via ALIX/syntenin/syndecan-4 required for completion of cytokinesis.Nat Commun. 2020 Apr 22;11(1):1941. doi: 10.1038/s41467-020-15205-z. Nat Commun. 2020. PMID: 32321914 Free PMC article.
-
Arabidopsis CaLB1 undergoes phase separation with the ESCRT protein ALIX and modulates autophagosome maturation.Nat Commun. 2024 Jun 19;15(1):5188. doi: 10.1038/s41467-024-49485-6. Nat Commun. 2024. PMID: 38898014 Free PMC article.
References
-
- Bache K.G., Stuffers S., Malerød L., Slagsvold T., Raiborg C., Lechardeur D., Wälchli S., Lukacs G.L., Brech A., and Stenmark H.. 2006. The ESCRT-III subunit hVps24 is required for degradation but not silencing of the epidermal growth factor receptor. Mol. Biol. Cell. 17:2513–2523. 10.1091/mbc.E05-10-0915 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

